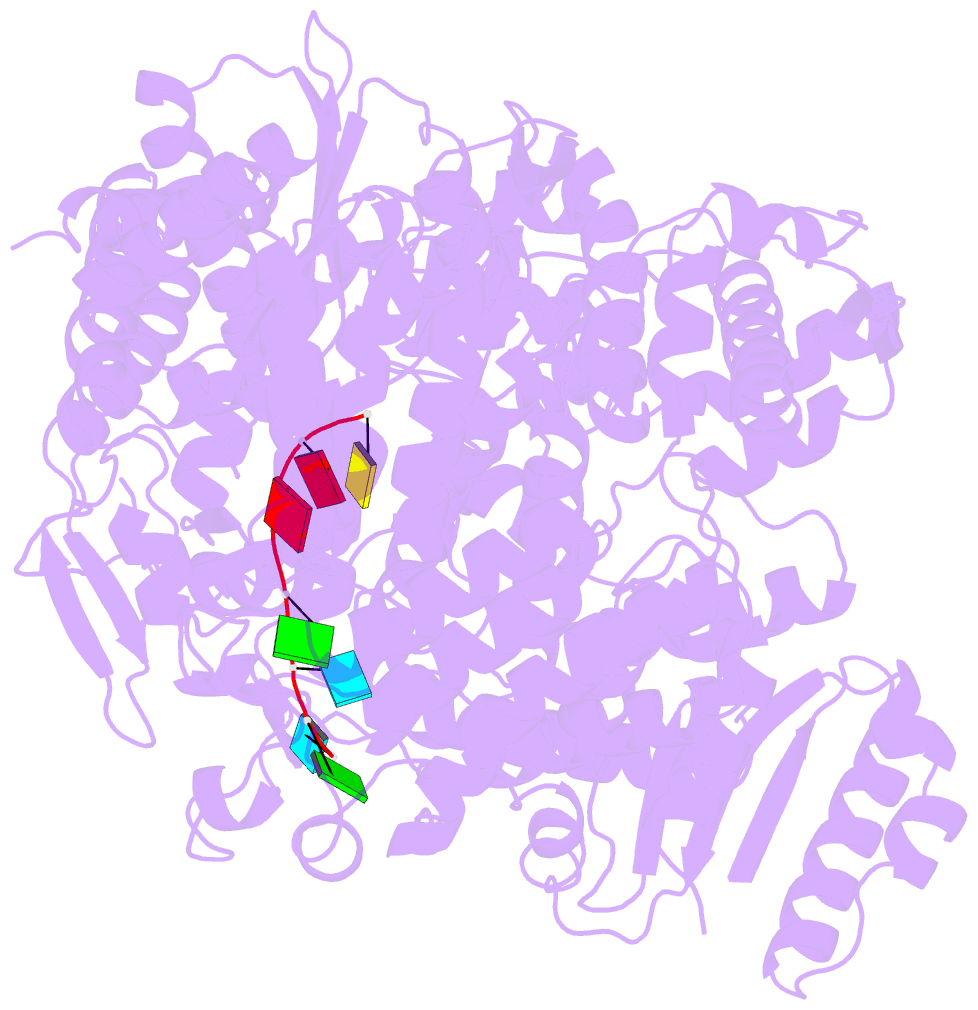
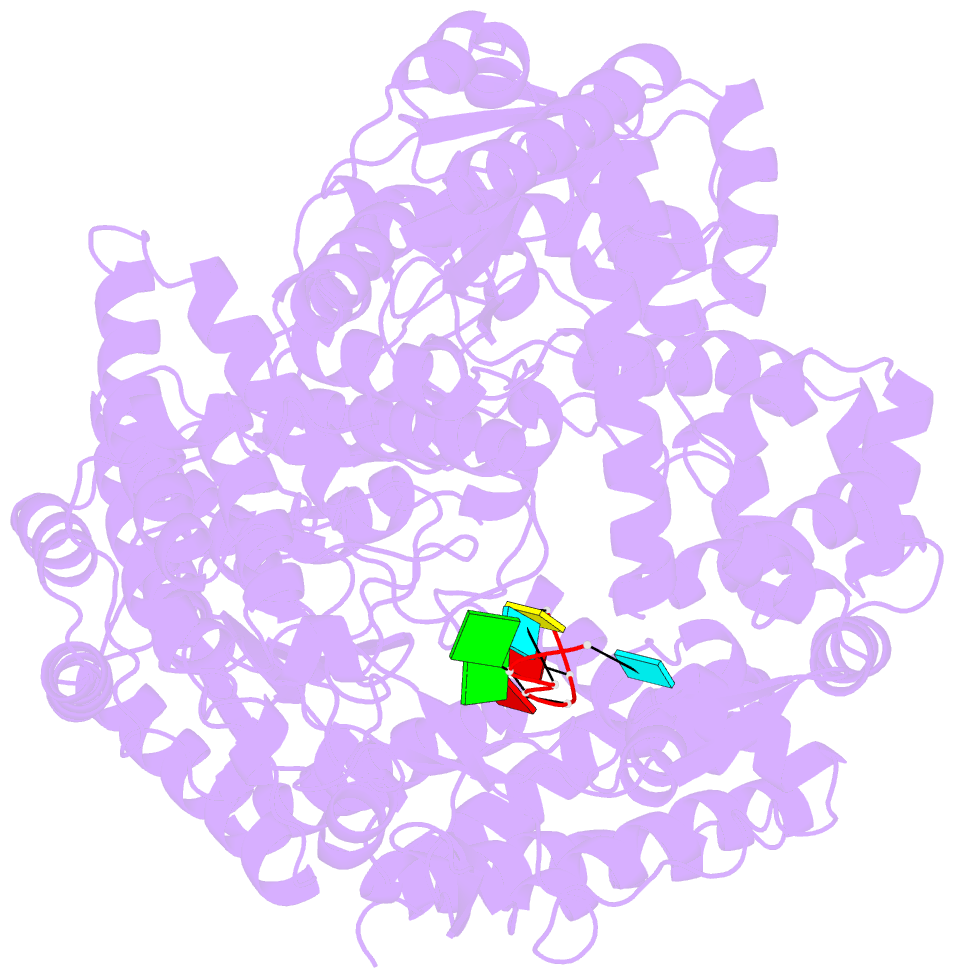
Summary information and primary citation
- PDB-id
- 2r7t; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-RNA
- Method
- X-ray (3.0 Å)
- Summary
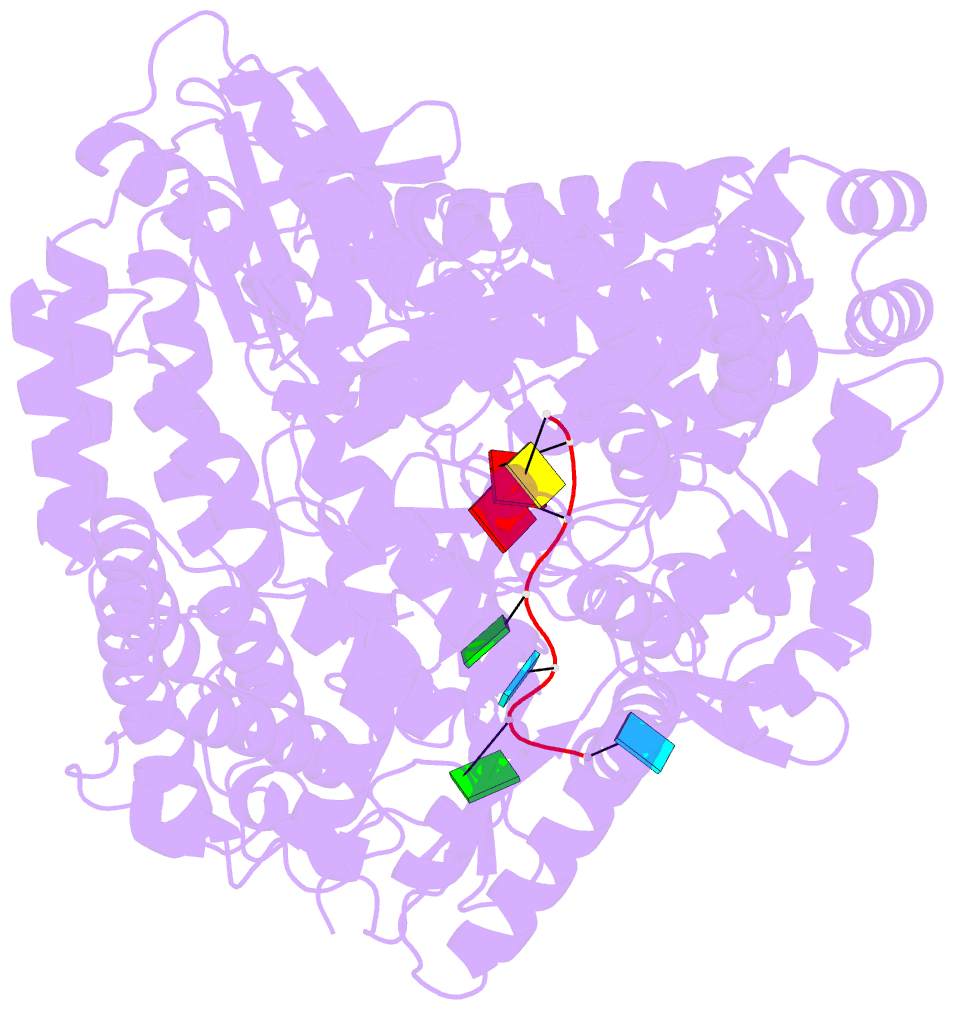
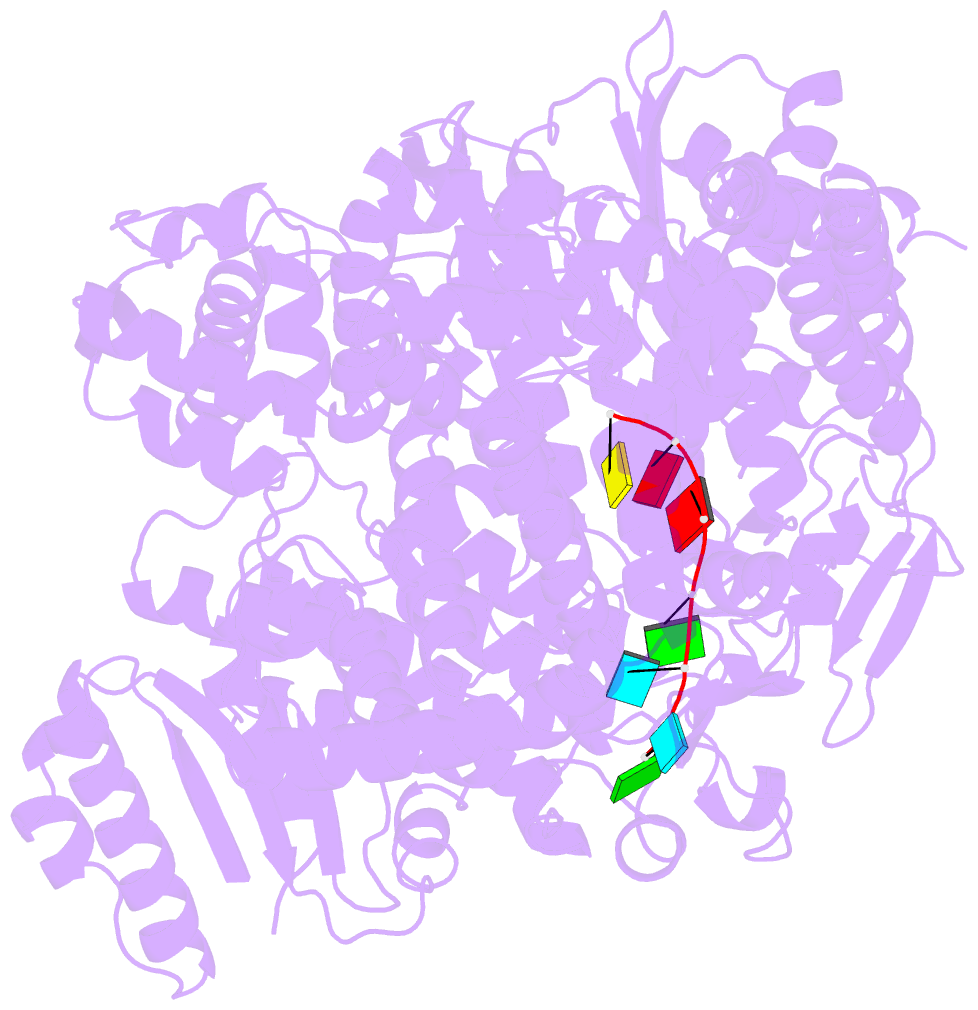
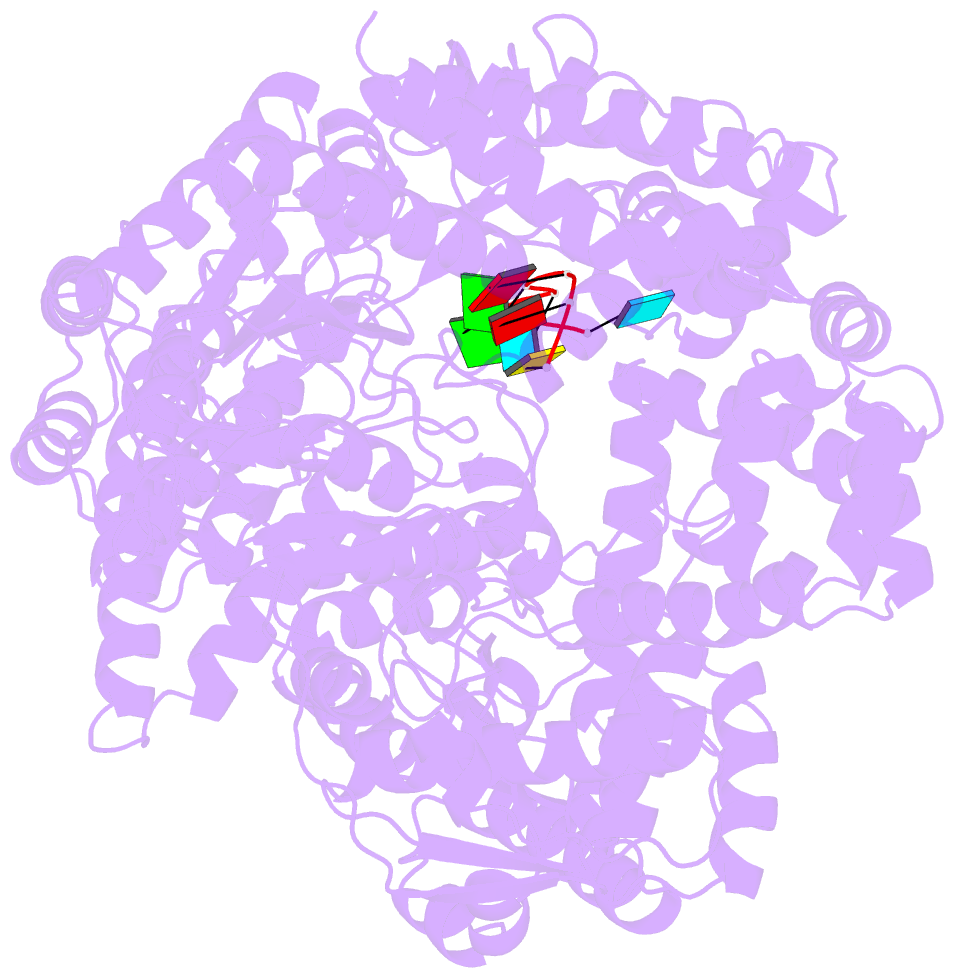
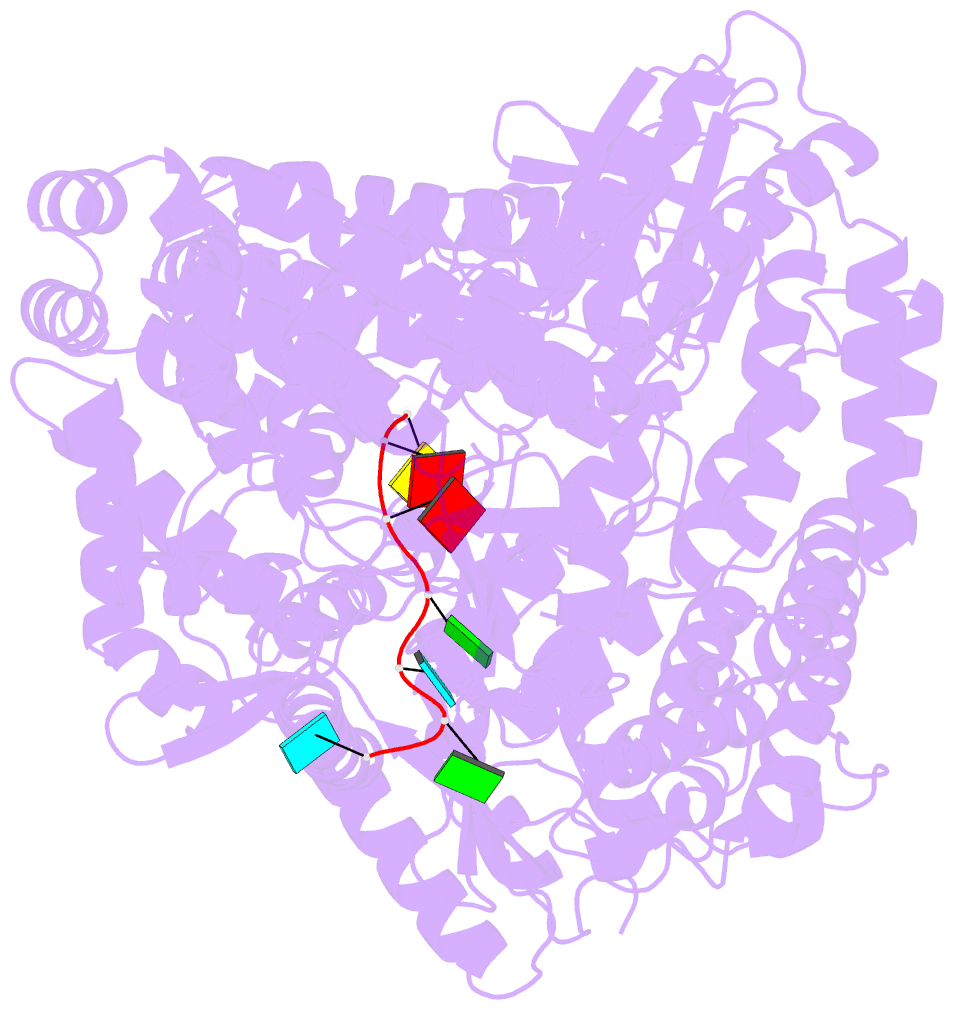
- Crystal structure of rotavirus sa11 vp1-RNA (ugugaacc) complex
- Reference
- Lu X, McDonald SM, Tortorici MA, Tao YJ, Vasquez-Del Carpio R, Nibert ML, Patton JT, Harrison SC (2008): "Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1." Structure, 16, 1678-1688. doi: 10.1016/j.str.2008.09.006.
- Abstract
- Rotavirus RNA-dependent RNA polymerase VP1 catalyzes RNA synthesis within a subviral particle. This activity depends on core shell protein VP2. A conserved sequence at the 3' end of plus-strand RNA templates is important for polymerase association and genome replication. We have determined the structure of VP1 at 2.9 A resolution, as apoenzyme and in complex with RNA. The cage-like enzyme is similar to reovirus lambda3, with four tunnels leading to or from a central, catalytic cavity. A distinguishing characteristic of VP1 is specific recognition, by conserved features of the template-entry channel, of four bases, UGUG, in the conserved 3' sequence. Well-defined interactions with these bases position the RNA so that its 3' end overshoots the initiating register, producing a stable but catalytically inactive complex. We propose that specific 3' end recognition selects rotavirus RNA for packaging and that VP2 activates the autoinhibited VP1/RNA complex to coordinate packaging and genome replication.