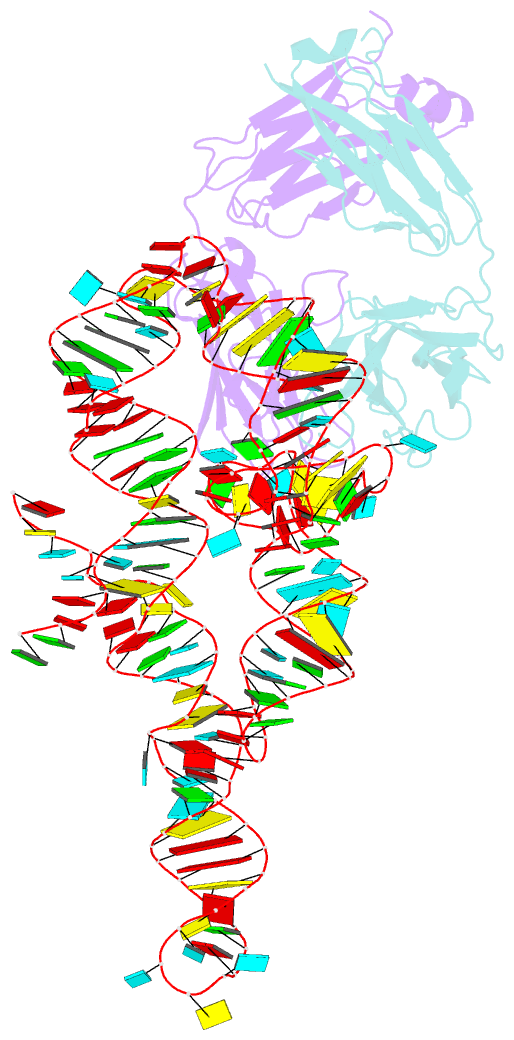
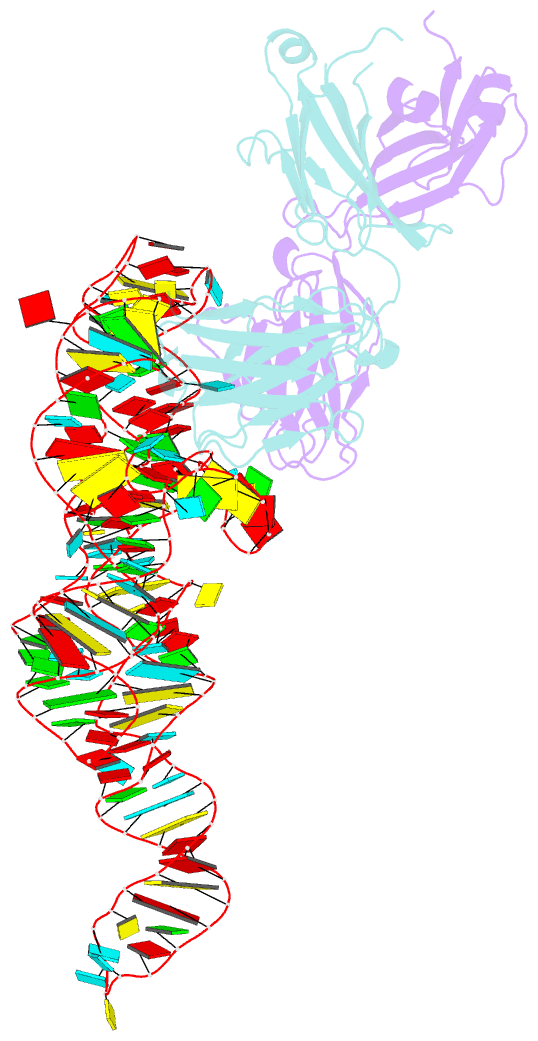
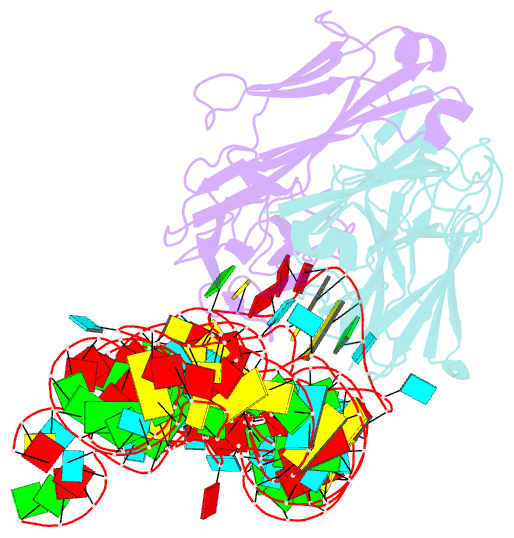
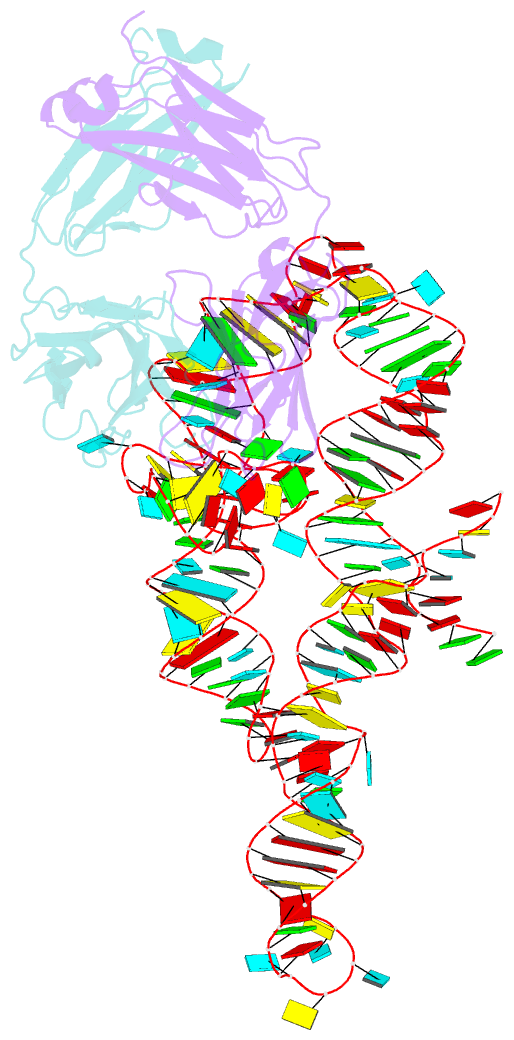
Summary information and primary citation
- PDB-id
- 2r8s; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- immune system-RNA
- Method
- X-ray (1.95 Å)
- Summary
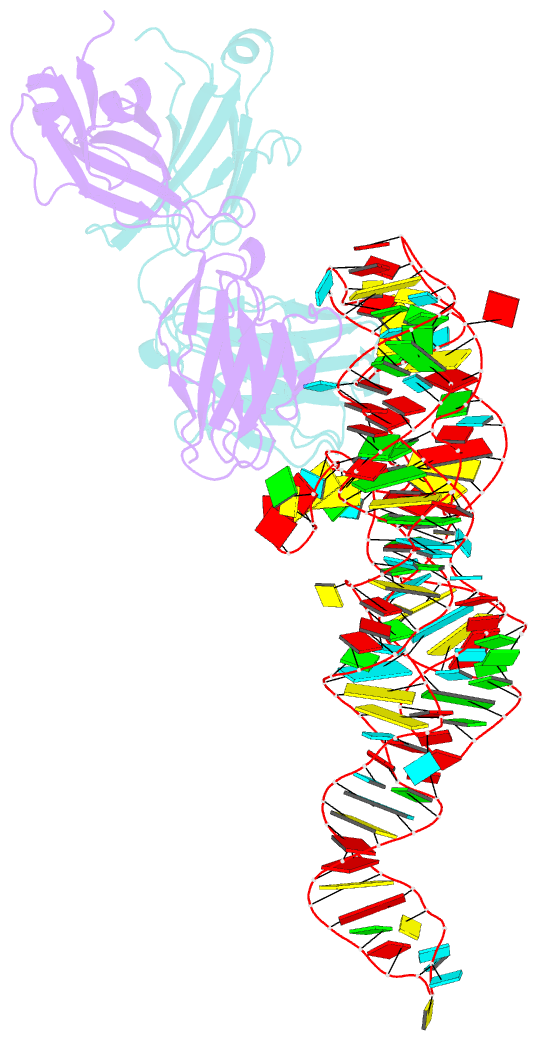
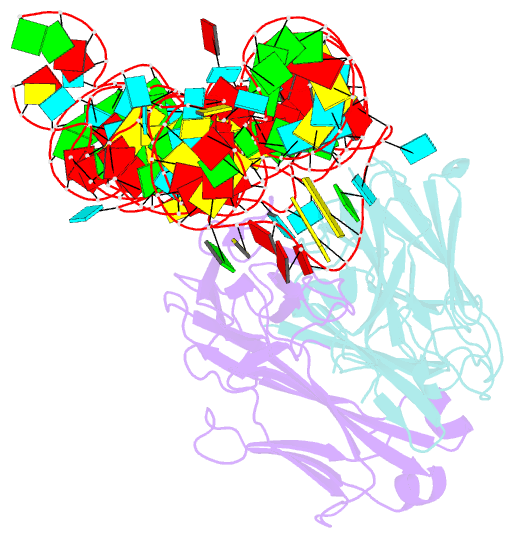
- High resolution structure of a specific synthetic fab bound to p4-p6 RNA ribozyme domain
- Reference
- Ye JD, Tereshko V, Frederiksen JK, Koide A, Fellouse FA, Sidhu SS, Koide S, Kossiakoff AA, Piccirilli JA (2008): "Synthetic antibodies for specific recognition and crystallization of structured RNA." Proc.Natl.Acad.Sci.Usa, 105, 82-87. doi: 10.1073/pnas.0709082105.
- Abstract
- Antibodies that bind protein antigens are indispensable in biochemical research and modern medicine. However, knowledge of RNA-binding antibodies and their application in the ever-growing RNA field is lacking. Here we have developed a robust approach using a synthetic phage-display library to select specific antigen-binding fragments (Fabs) targeting a large functional RNA. We have solved the crystal structure of the first Fab-RNA complex at 1.95 A. Capability in phasing and crystal contact formation suggests that the Fab provides a potentially valuable crystal chaperone for RNA. The crystal structure reveals that the Fab achieves specific RNA binding on a shallow surface with complementarity-determining region (CDR) sequence diversity, length variability, and main-chain conformational plasticity. The Fab-RNA interface also differs significantly from Fab-protein interfaces in amino acid composition and light-chain participation. These findings yield valuable insights for engineering of Fabs as RNA-binding modules and facilitate further development of Fabs as possible therapeutic drugs and biochemical tools to explore RNA biology.