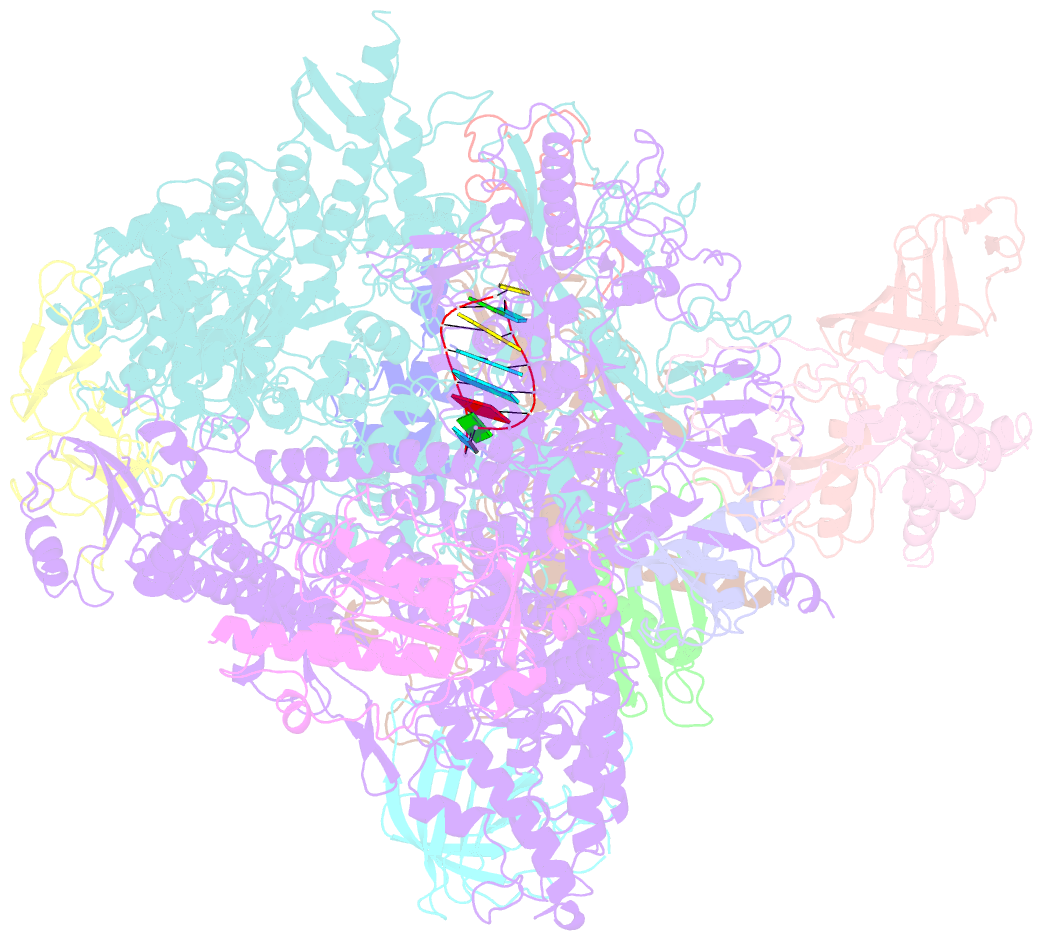
Summary information and primary citation
- PDB-id
- 2r93; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-RNA
- Method
- X-ray (4.0 Å)
- Summary
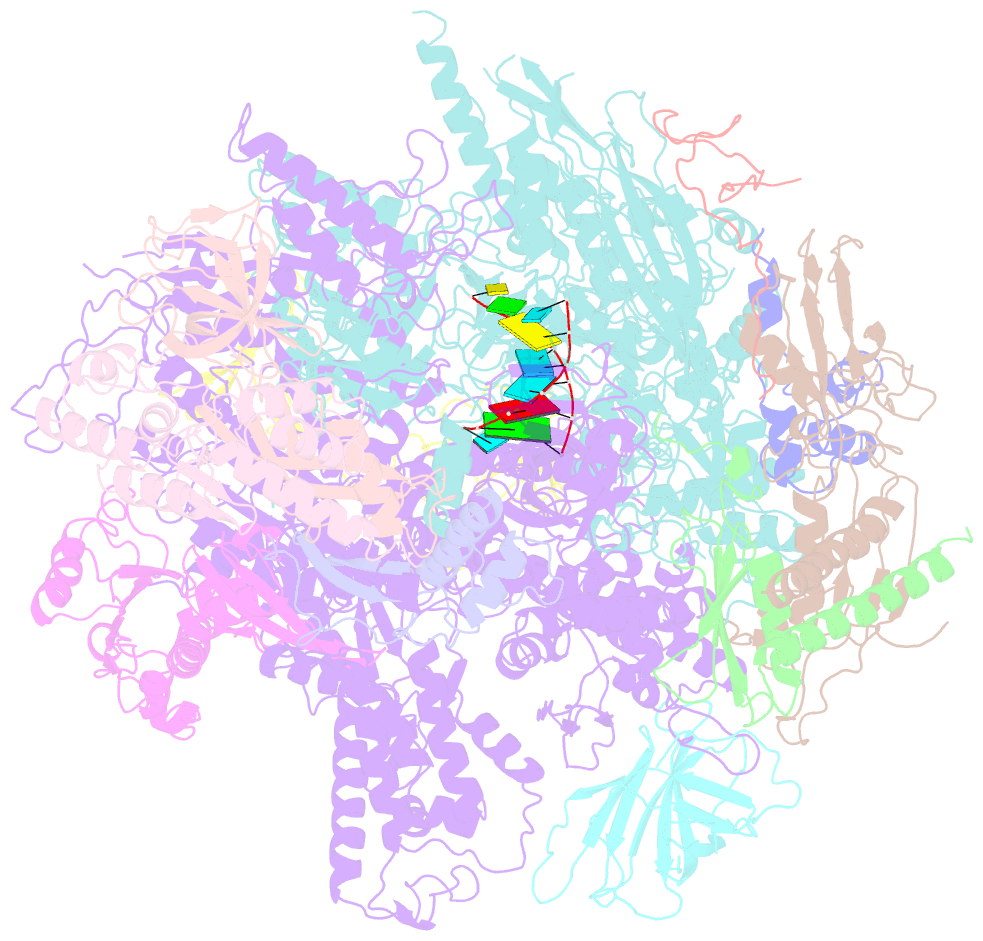
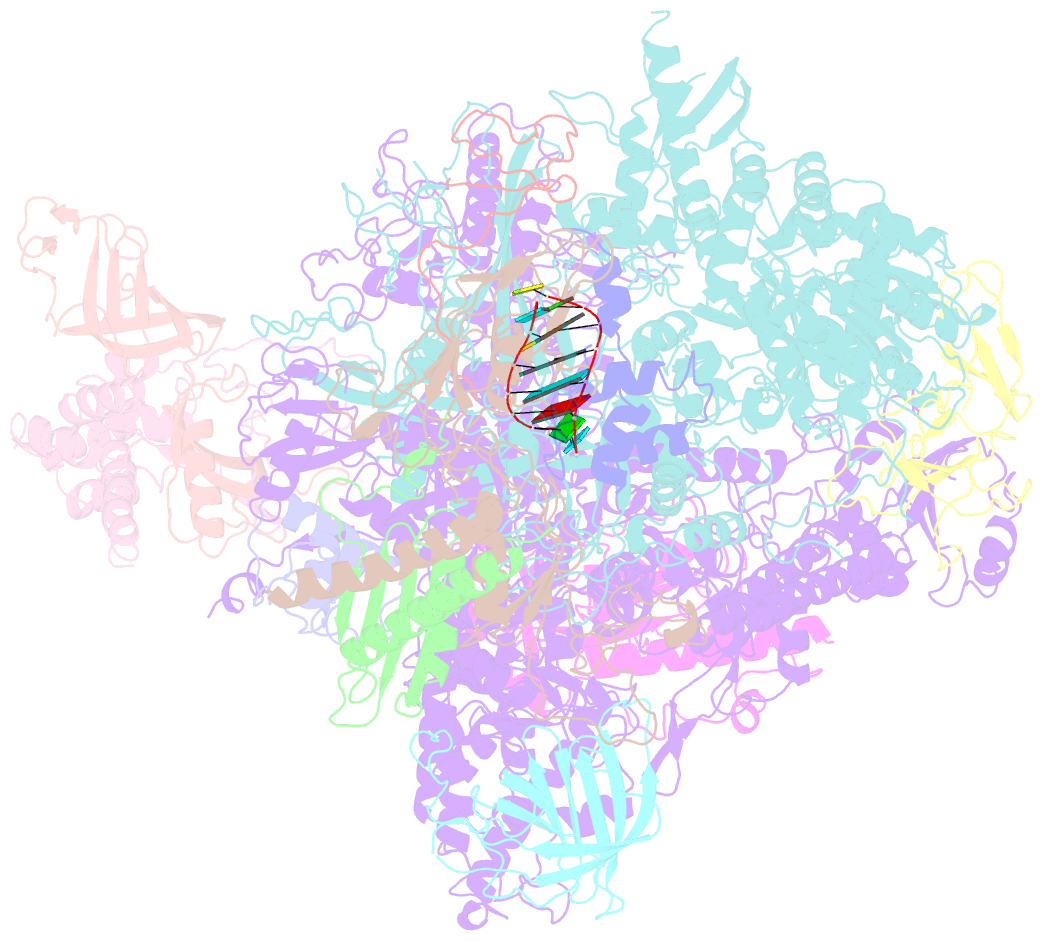
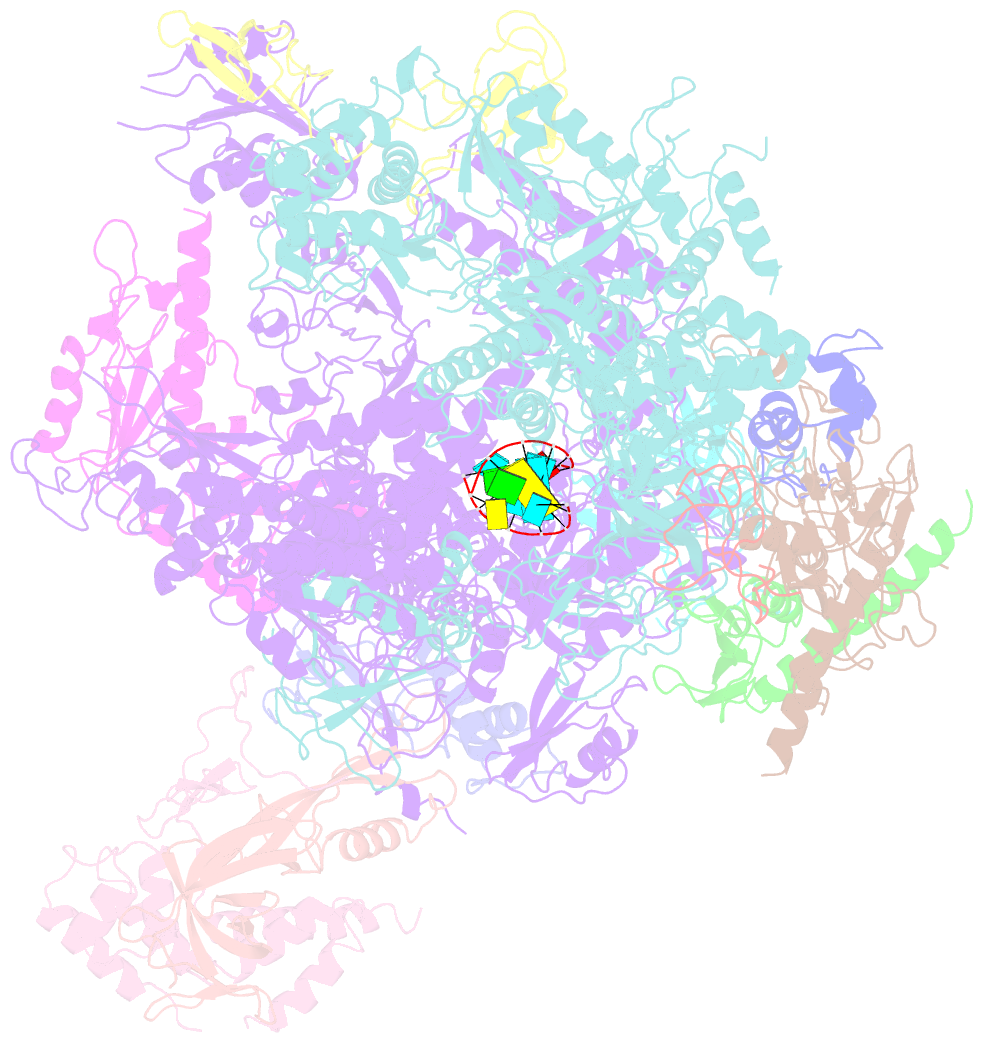
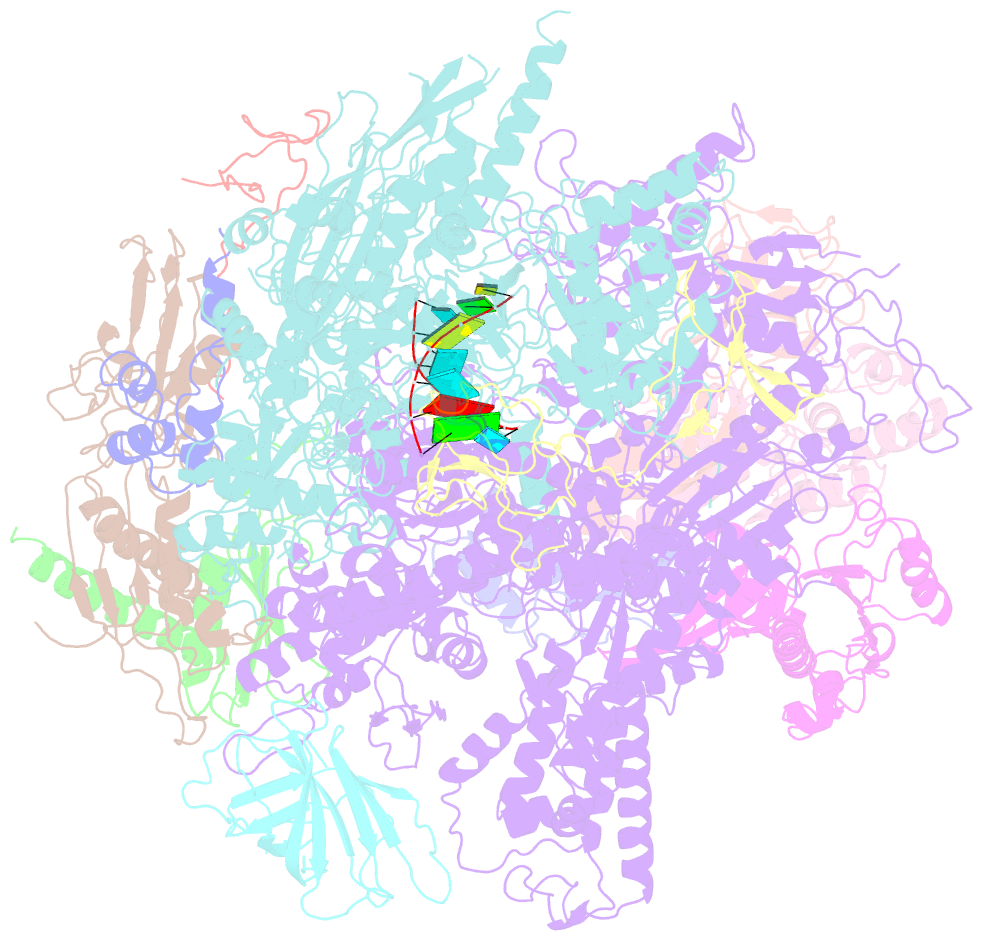
- Elongation complex of RNA polymerase ii with a hepatitis delta virus-derived RNA stem loop
- Reference
- Lehmann E, Brueckner F, Cramer P (2007): "Molecular basis of RNA-dependent RNA polymerase II activity." Nature, 450, 445-449. doi: 10.1038/nature06290.
- Abstract
- RNA polymerase (Pol) II catalyses DNA-dependent RNA synthesis during gene transcription. There is, however, evidence that Pol II also possesses RNA-dependent RNA polymerase (RdRP) activity. Pol II can use a homopolymeric RNA template, can extend RNA by several nucleotides in the absence of DNA, and has been implicated in the replication of the RNA genomes of hepatitis delta virus (HDV) and plant viroids. Here we show the intrinsic RdRP activity of Pol II with only pure polymerase, an RNA template-product scaffold and nucleoside triphosphates (NTPs). Crystallography reveals the template-product duplex in the site occupied by the DNA-RNA hybrid during transcription. RdRP activity resides at the active site used during transcription, but it is slower and less processive than DNA-dependent activity. RdRP activity is also obtained with part of the HDV antigenome. The complex of transcription factor IIS (TFIIS) with Pol II can cleave one HDV strand, create a reactive stem-loop in the hybrid site, and extend the new RNA 3' end. Short RNA stem-loops with a 5' extension suffice for activity, but their growth to a critical length apparently impairs processivity. The RdRP activity of Pol II provides a missing link in molecular evolution, because it suggests that Pol II evolved from an ancient replicase that duplicated RNA genomes.