Summary information and primary citation
- PDB-id
- 2rfk; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- isomerase-RNA
- Method
- X-ray (2.87 Å)
- Summary
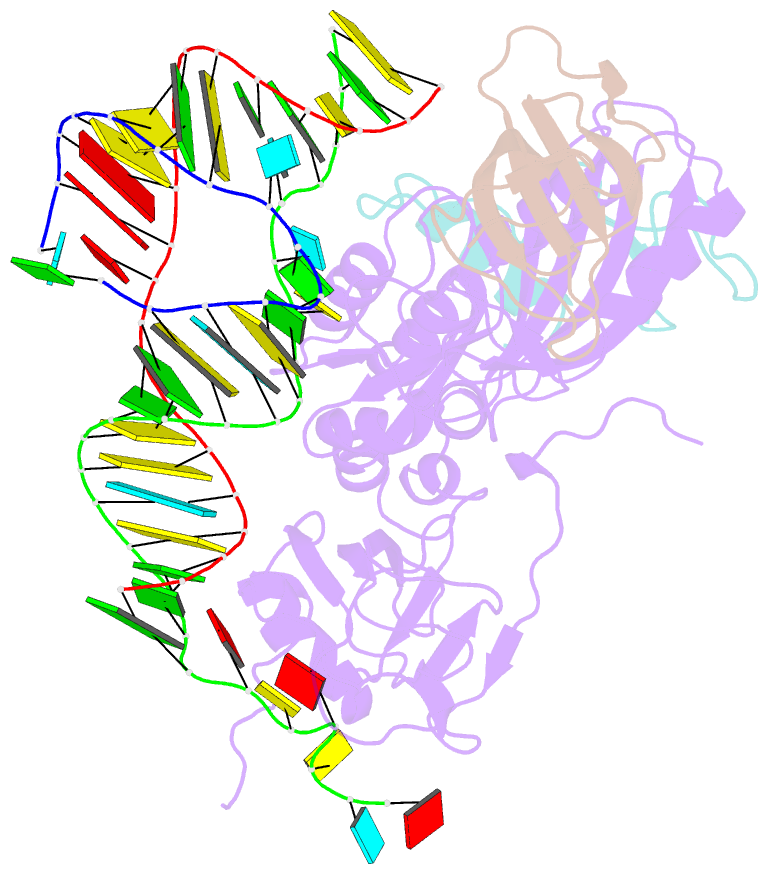
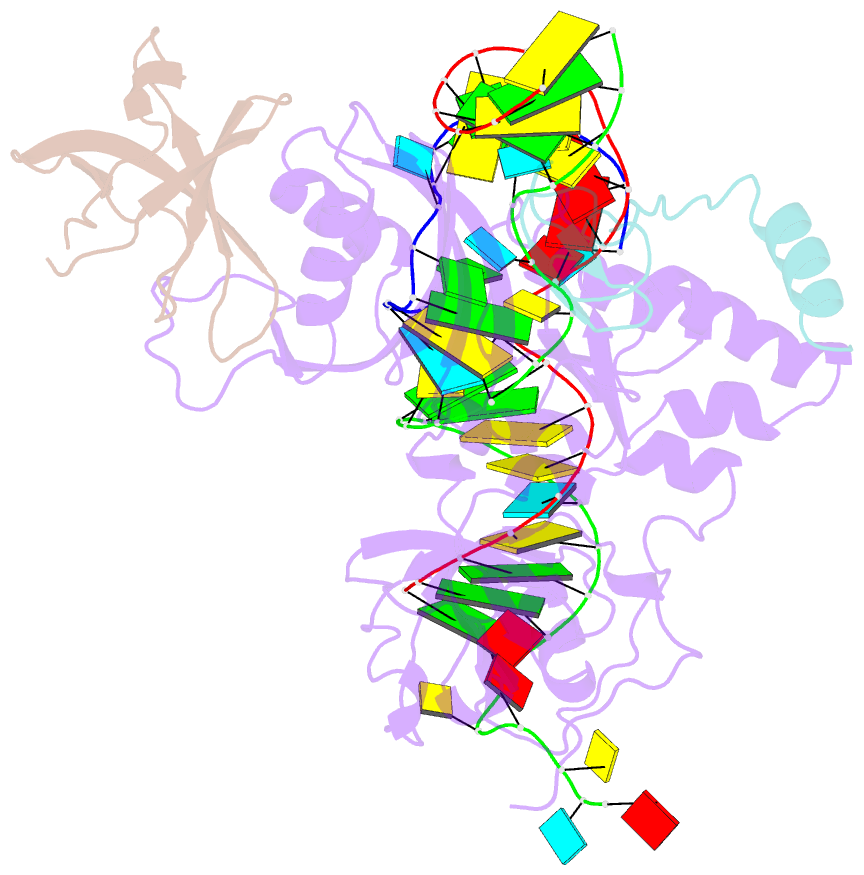
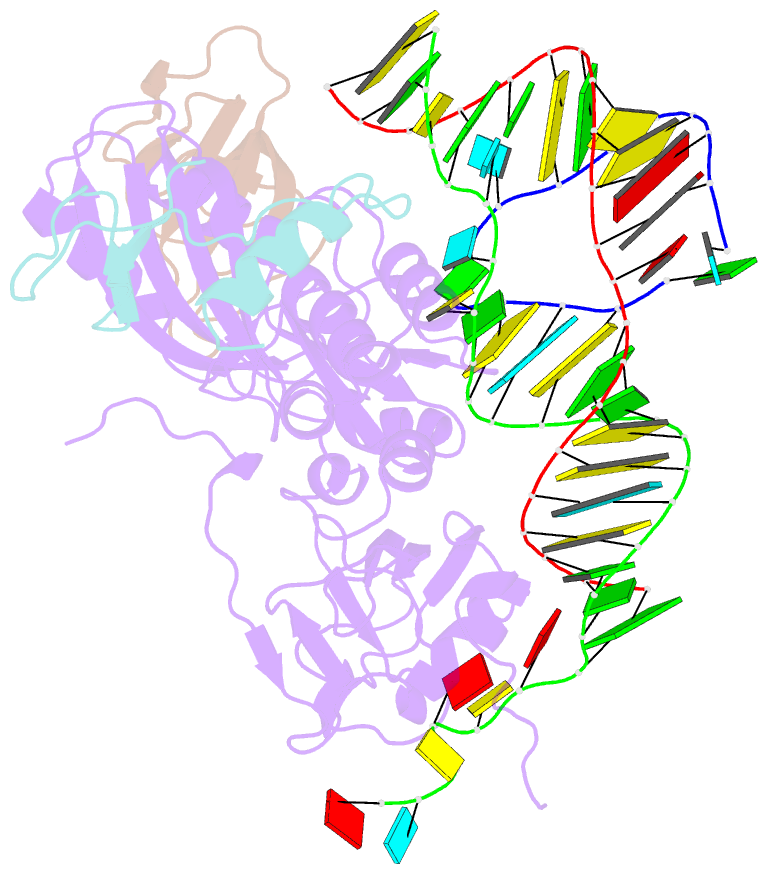
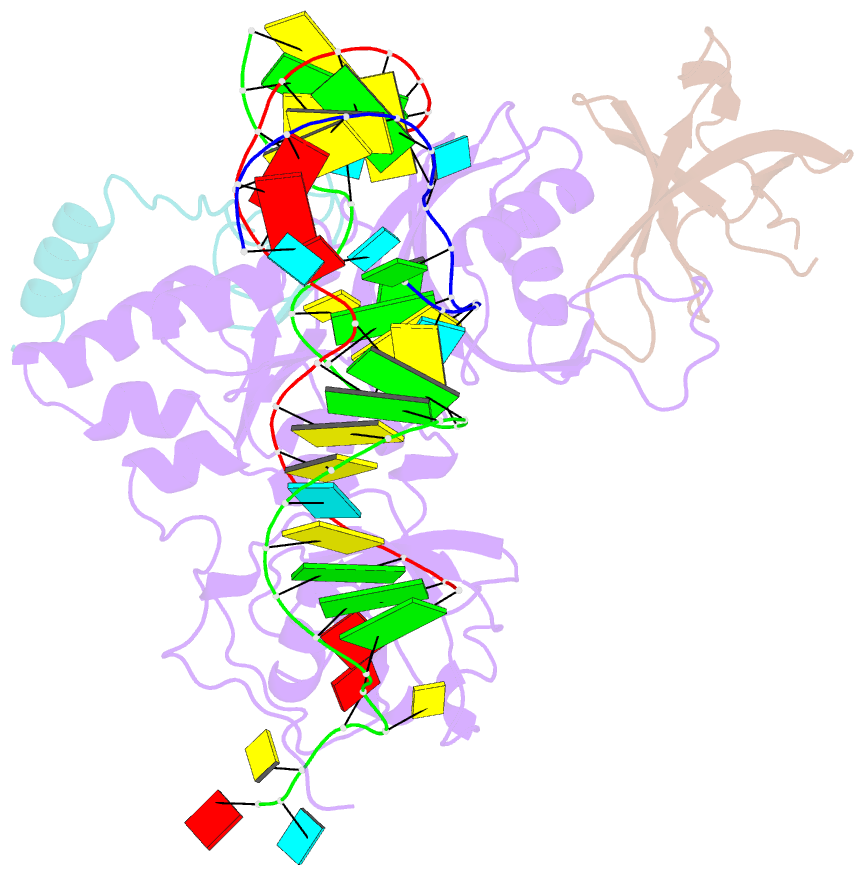
- Substrate RNA positioning in the archaeal h-aca ribonucleoprotein complex
- Reference
- Liang B, Xue S, Terns RM, Terns MP, Li H (2007): "Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex." Nat.Struct.Mol.Biol., 14, 1189-1195. doi: 10.1038/nsmb1336.
- Abstract
- The most complex RNA pseudouridylases are H/ACA ribonucleoprotein particles, which use a guide RNA for substrate capture and four proteins (Cbf5, Nop10, Gar1 and L7Ae/NHP2) for substrate modification. Here we report the three-dimensional structure of a catalytically deficient archaeal enzyme complex (including the guide RNA and three of the four essential proteins) bound to a substrate RNA. Extensive interactions of Cbf5 with one guide-substrate helix and a guide RNA stem shape the forked guide–substrate RNA complex structure and position the substrate in proximity of the Cbf5 catalytic center. Our structural and complementary fluorescence analyses also indicate that precise placement of the target uridine at the active site requires a conformation of the guide–substrate RNA duplex that is brought about by the previously identified concurrent interaction of the guide RNA with L7Ae and a composite Cbf5-Nop10 surface, and further identify a residue that is critical in this process.