Summary information and primary citation
- PDB-id
- 2tmv; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- virus-RNA
- Method
- fiber diffraction
- Summary
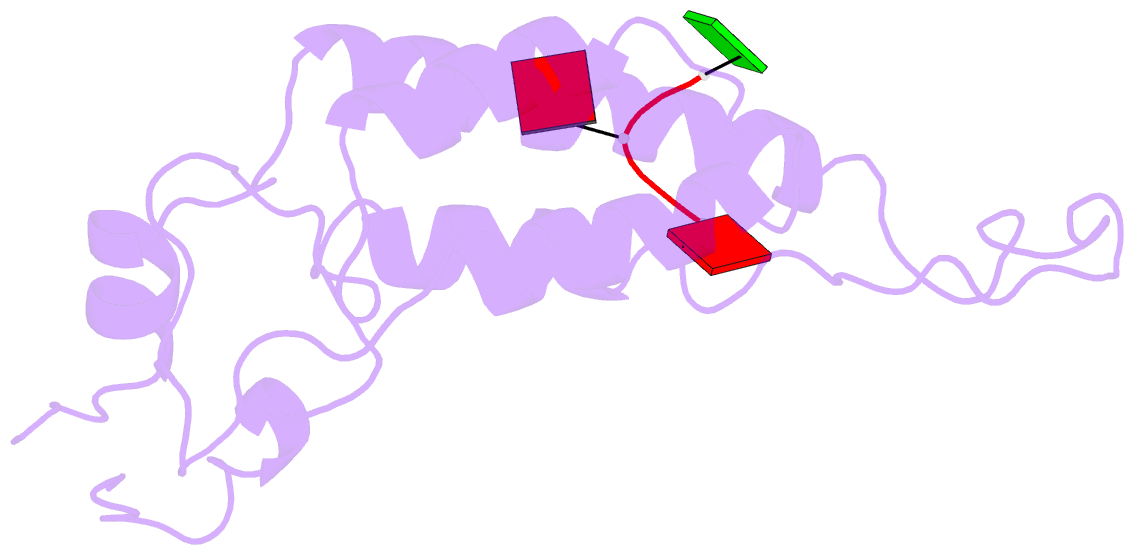
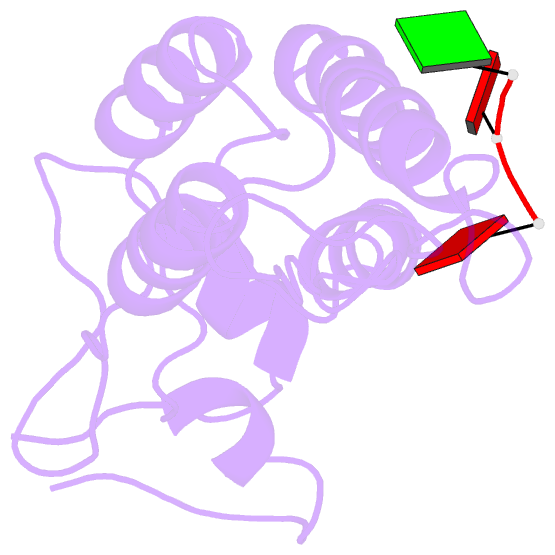
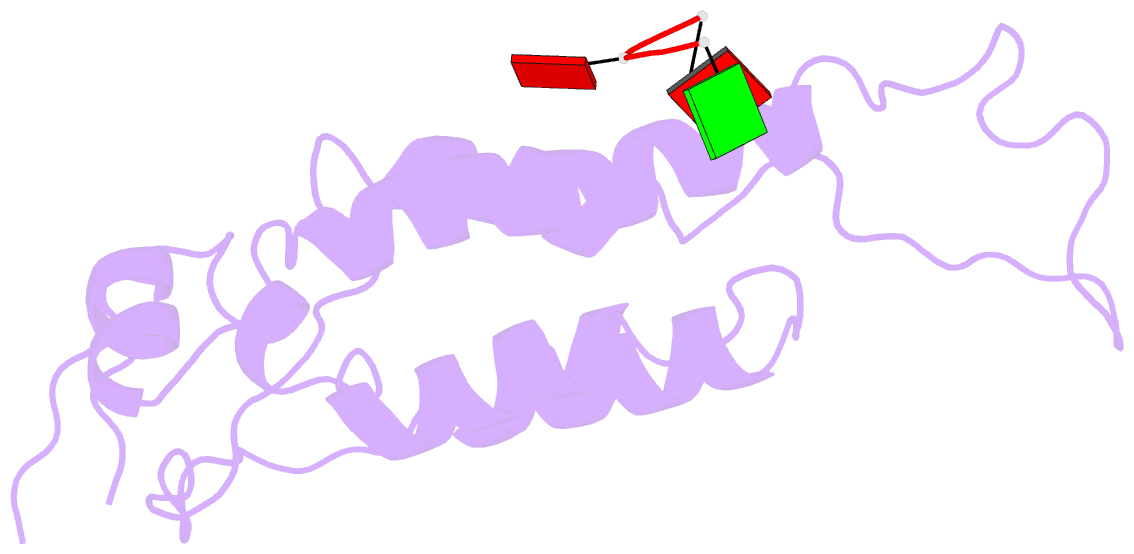
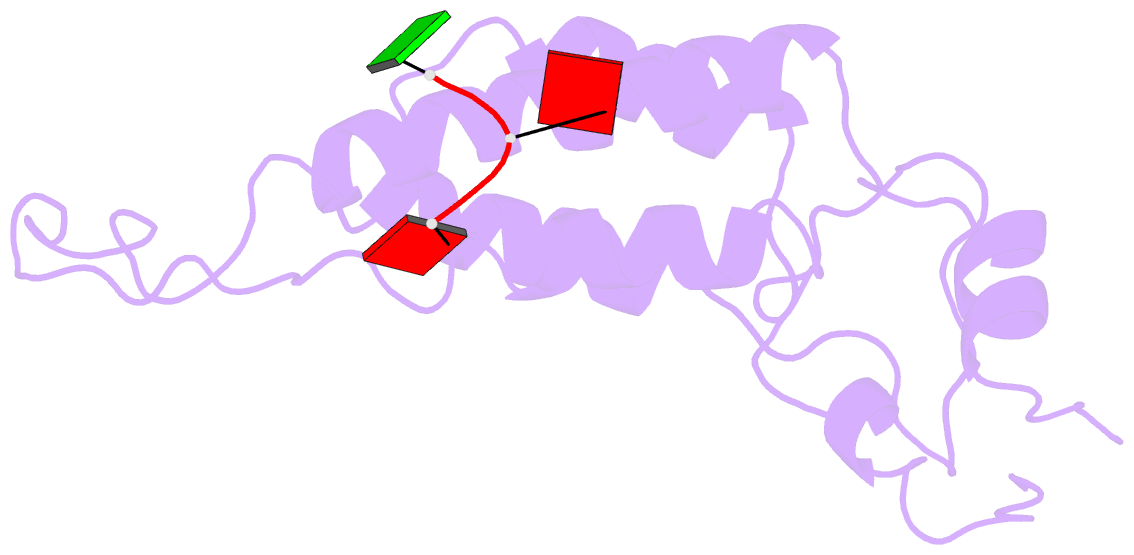
- Visualization of protein-nucleic acid interactions in a virus. refined structure of intact tobacco mosaic virus at 2.9 angstroms resolution by x-ray fiber diffraction
- Reference
- Namba K, Pattanayek R, Stubbs G (1989): "Visualization of protein-nucleic acid interactions in a virus. Refined structure of intact tobacco mosaic virus at 2.9 A resolution by X-ray fiber diffraction." J.Mol.Biol., 208, 307-325. doi: 10.1016/0022-2836(89)90391-4.
- Abstract
- The structure of tobacco mosaic virus (TMV) has been determined by fiber diffraction methods at 2.9 A resolution, and refined by restrained least-squares to an R-factor of 0.096. Protein-nucleic acid interactions are clearly visible. The final model contains all of the non-hydrogen atoms of the RNA and the protein, 71 water molecules, and two calcium-binding sites. Viral disassembly is driven by electrostatic repulsions between the charges in two carboxyl-carboxylate pairs and a phosphate-carboxylate pair. The phosphate-carboxylate pair and at least one of the carboxyl-carboxylate pairs appear to be calcium-binding sites. Nucleotide specificity, enabling TMV to recognize its own RNA by a repeating pattern of guanine residues, is provided by two guanine-specific hydrogen bonds in one of the three base-binding sites.