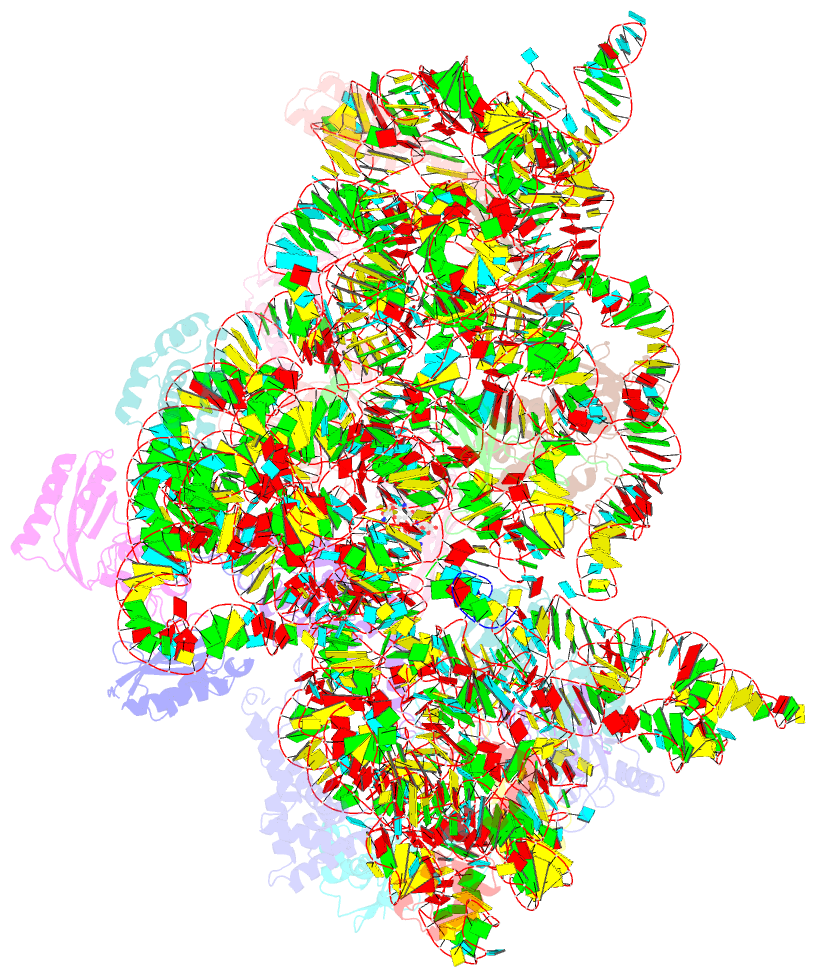
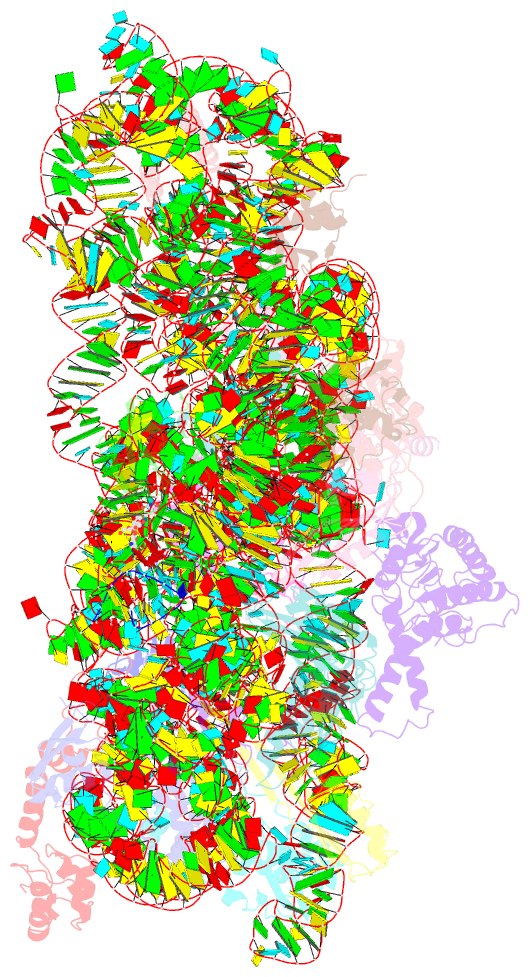
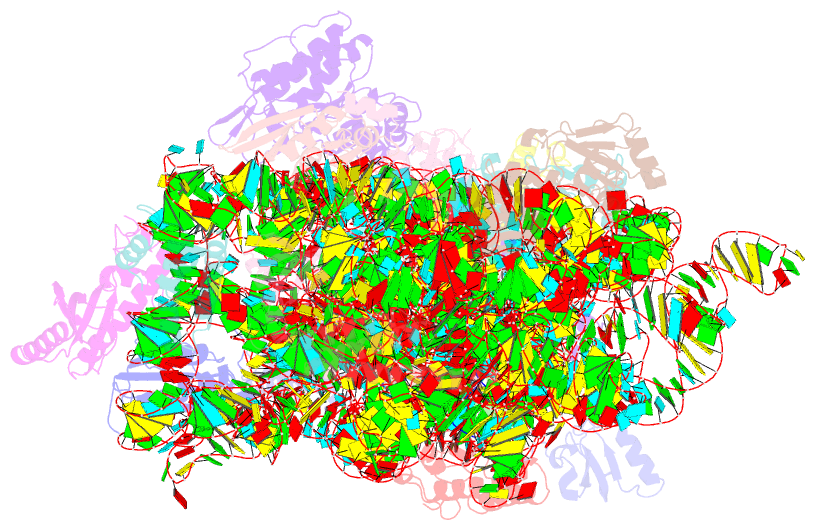
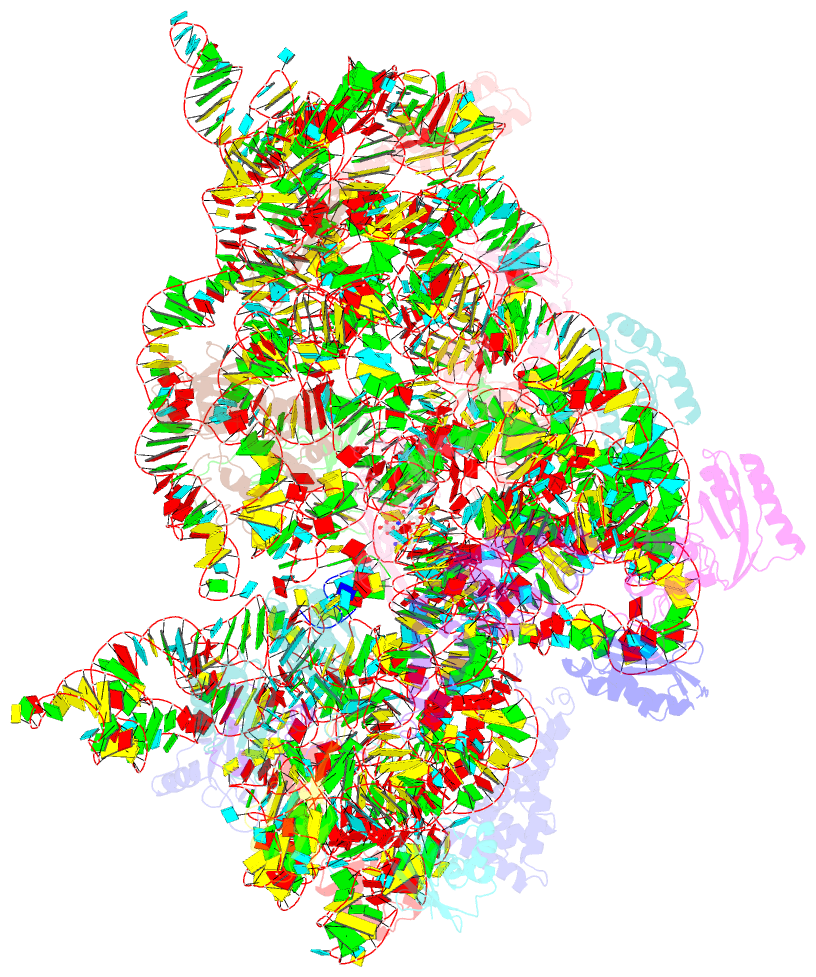
Summary information and primary citation
- PDB-id
- 2uub; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- X-ray (2.8 Å)
- Summary
- Structure of the thermus thermophilus 30s ribosomal subunit complexed with a valine-asl with cmo5u in position 34 bound to an mrna with a guu-codon in the a-site and paromomycin.
- Reference
- Weixlbaumer A, Murphy FV, Dziergowska A, Malkiewicz A, Vendeix FAP, Agris PF, Ramakrishnan V (2007): "Mechanism for Expanding the Decoding Capacity of Transfer Rnas by Modification of Uridines." Nat.Struct.Mol.Biol., 14, 498. doi: 10.1038/NSMB1242.
- Abstract
- One of the most prevalent base modifications involved in decoding is uridine 5-oxyacetic acid at the wobble position of tRNA. It has been known for several decades that this modification enables a single tRNA to decode all four codons in a degenerate codon box. We have determined structures of an anticodon stem-loop of tRNA(Val) containing the modified uridine with all four valine codons in the decoding site of the 30S ribosomal subunit. An intramolecular hydrogen bond involving the modification helps to prestructure the anticodon loop. We found unusual base pairs with the three noncomplementary codon bases, including a G.U base pair in standard Watson-Crick geometry, which presumably involves an enol form for the uridine. These structures suggest how a modification in the uridine at the wobble position can expand the decoding capability of a tRNA.