Summary information and primary citation
- PDB-id
- 2vnu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.3 Å)
- Summary
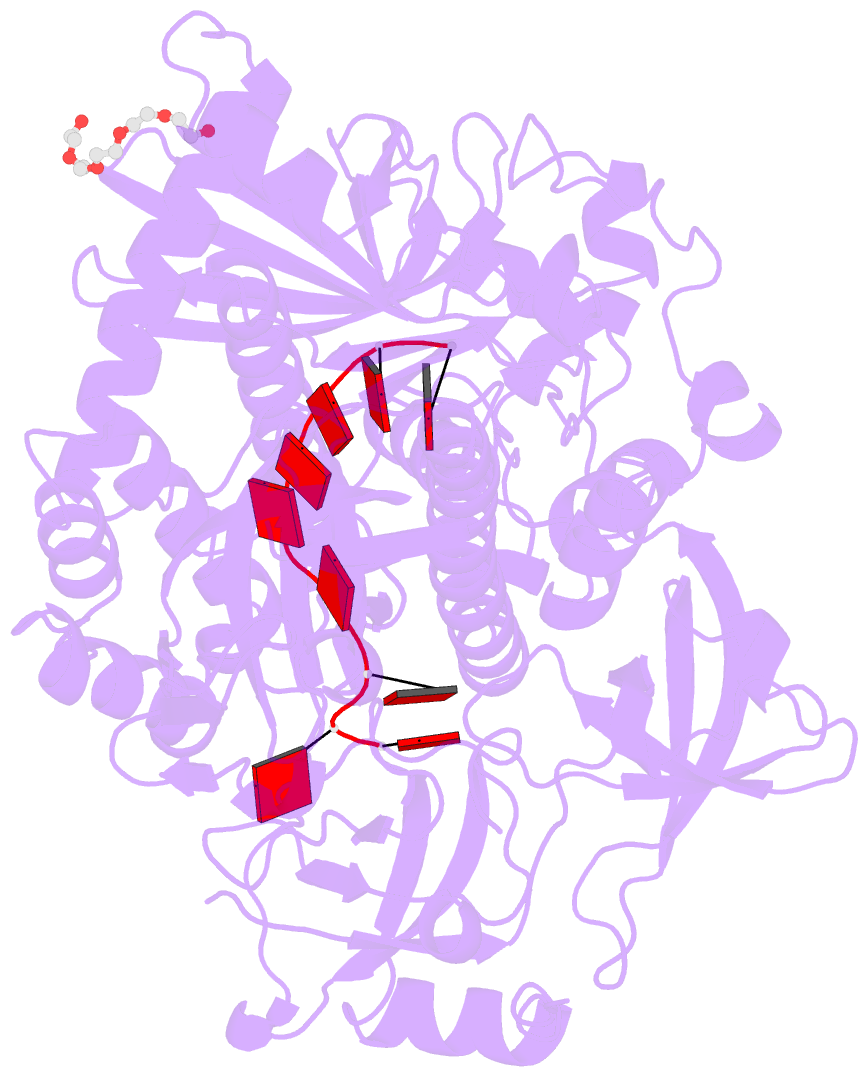
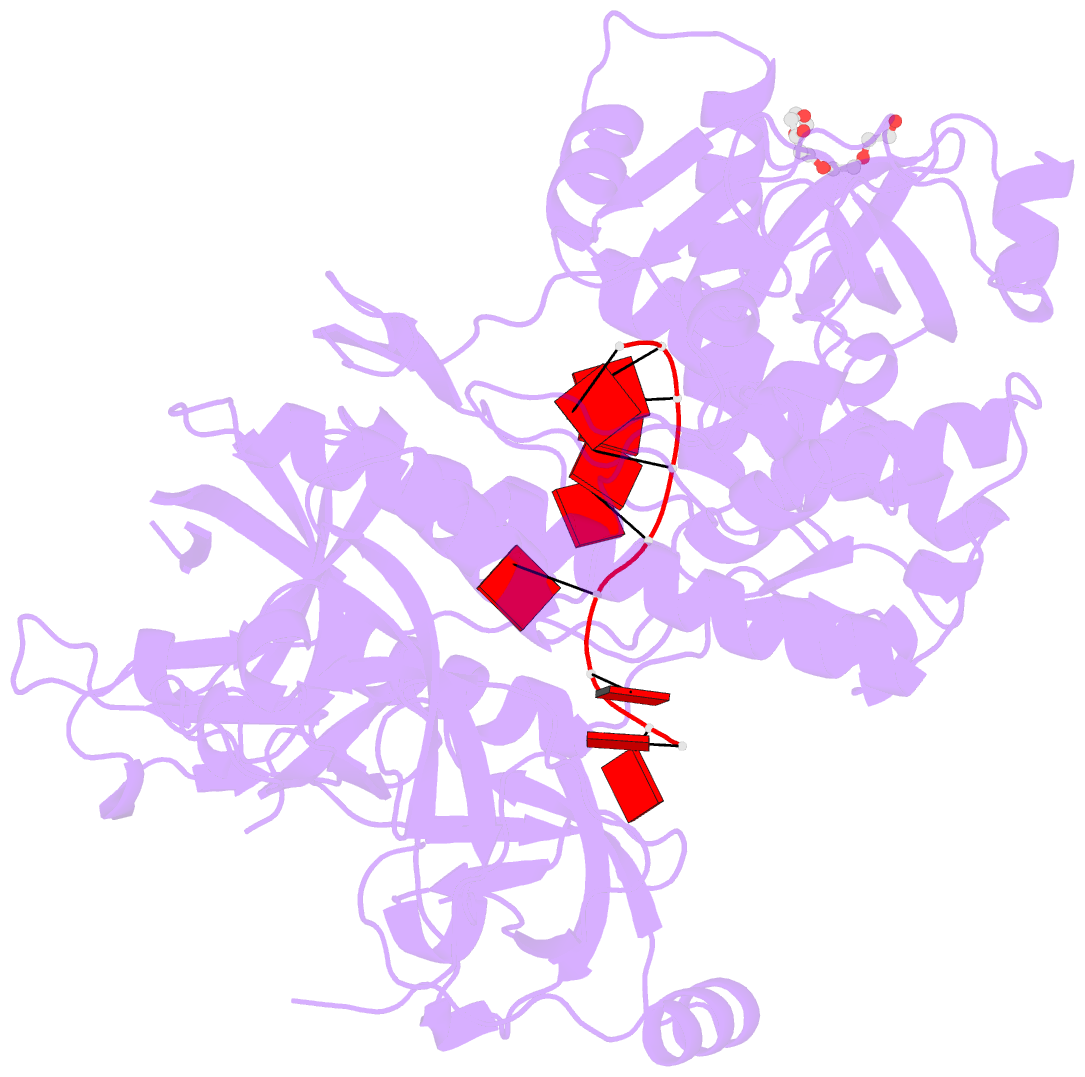
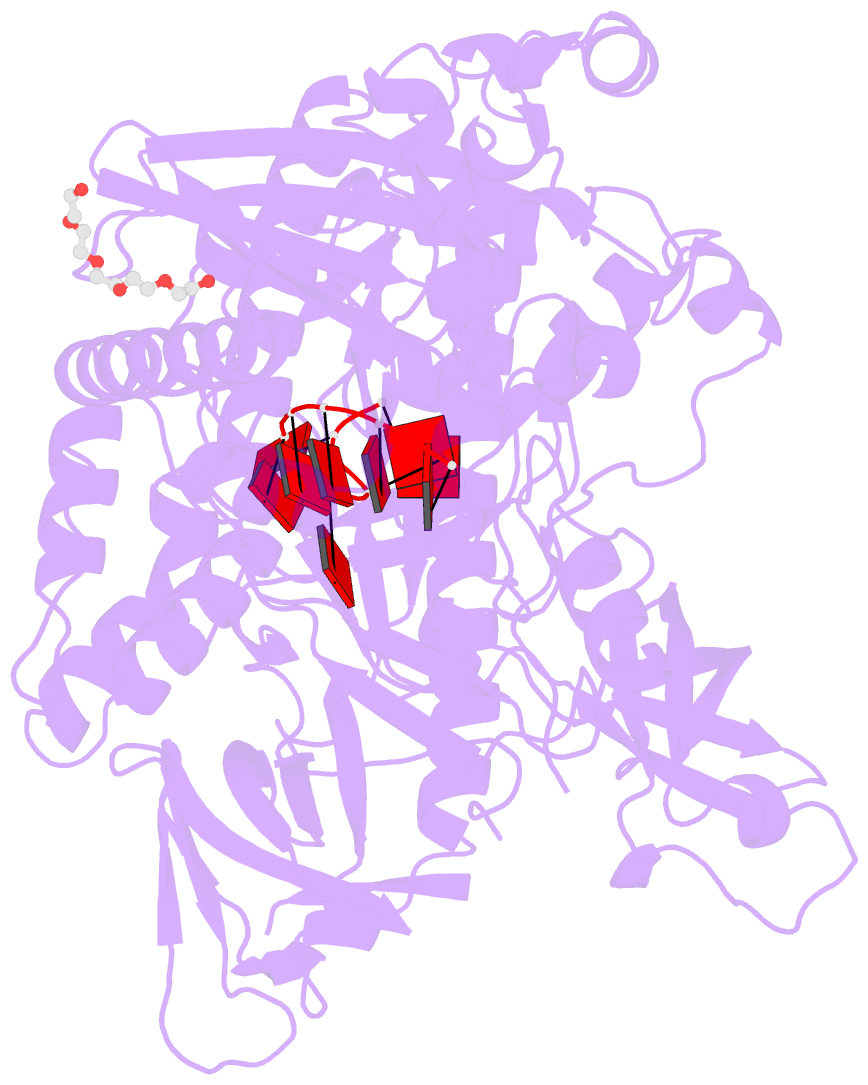
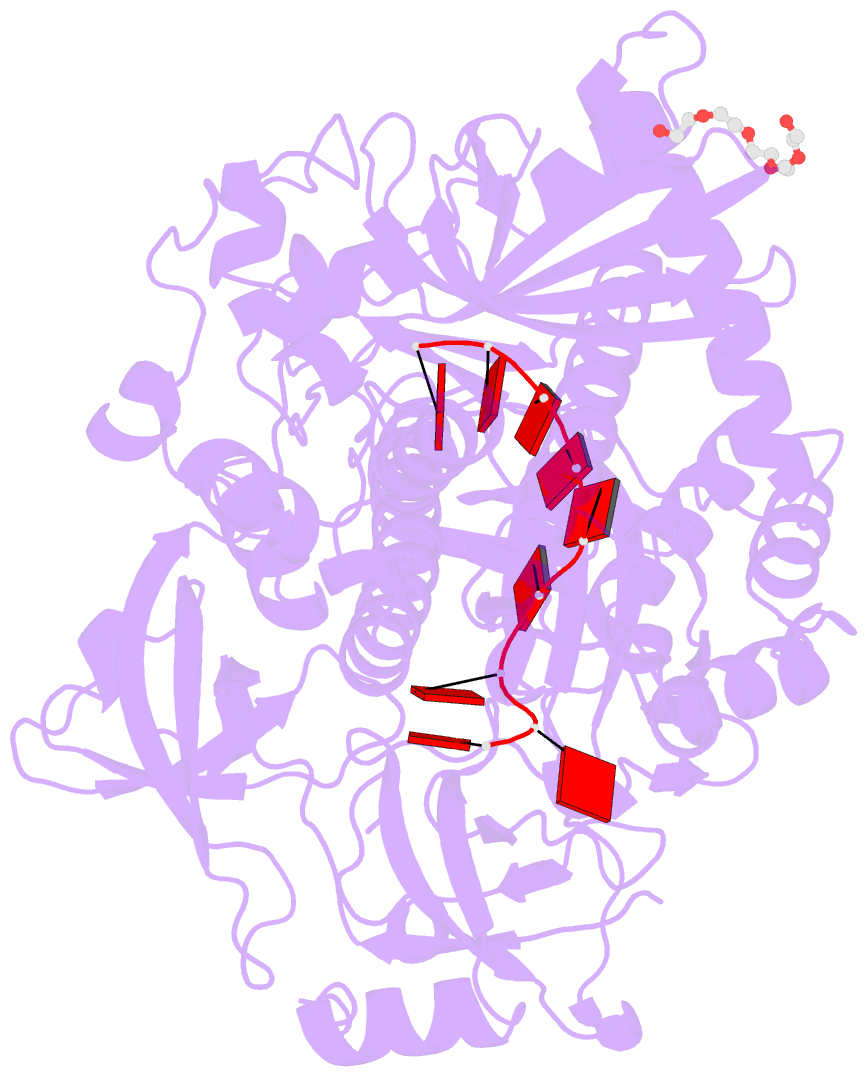
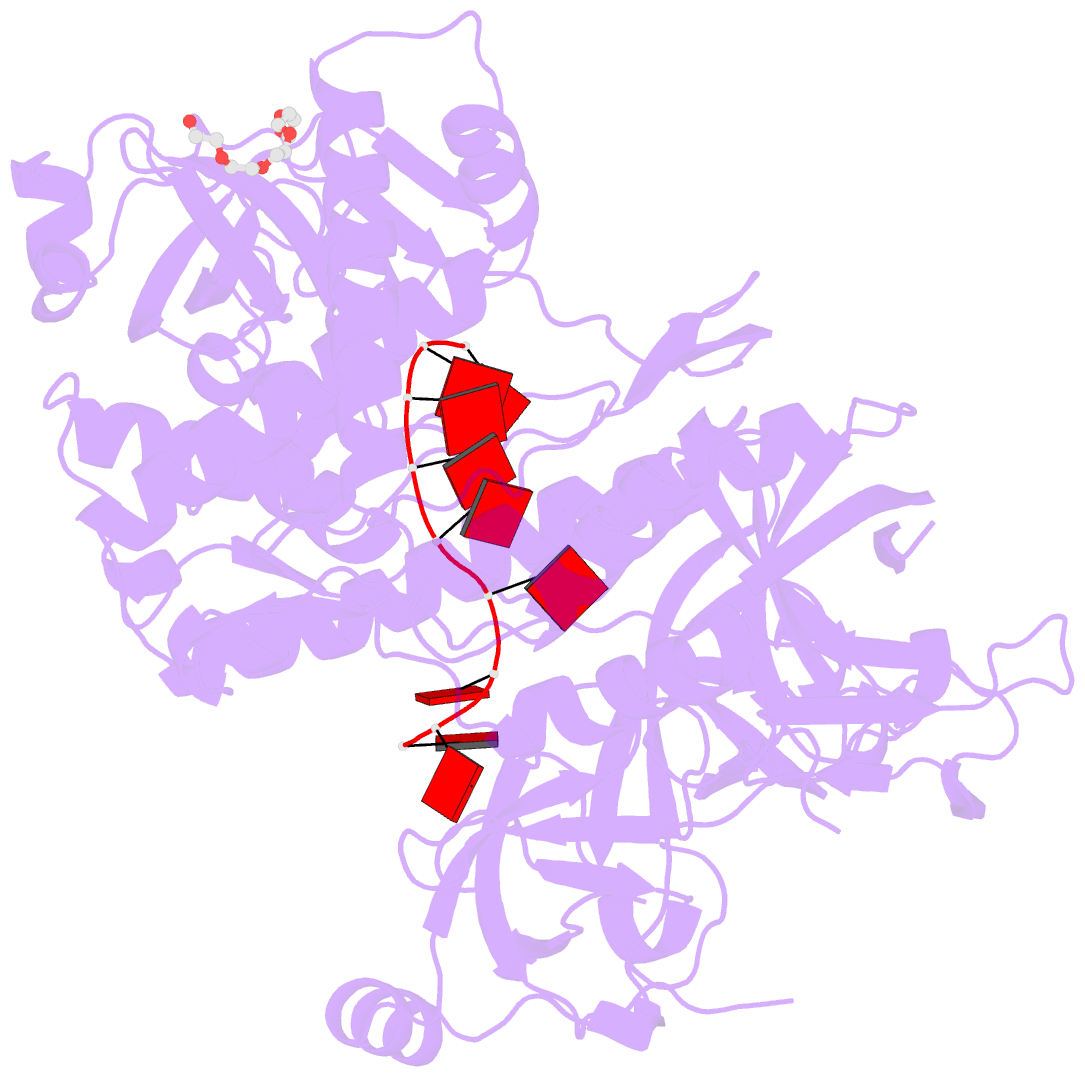
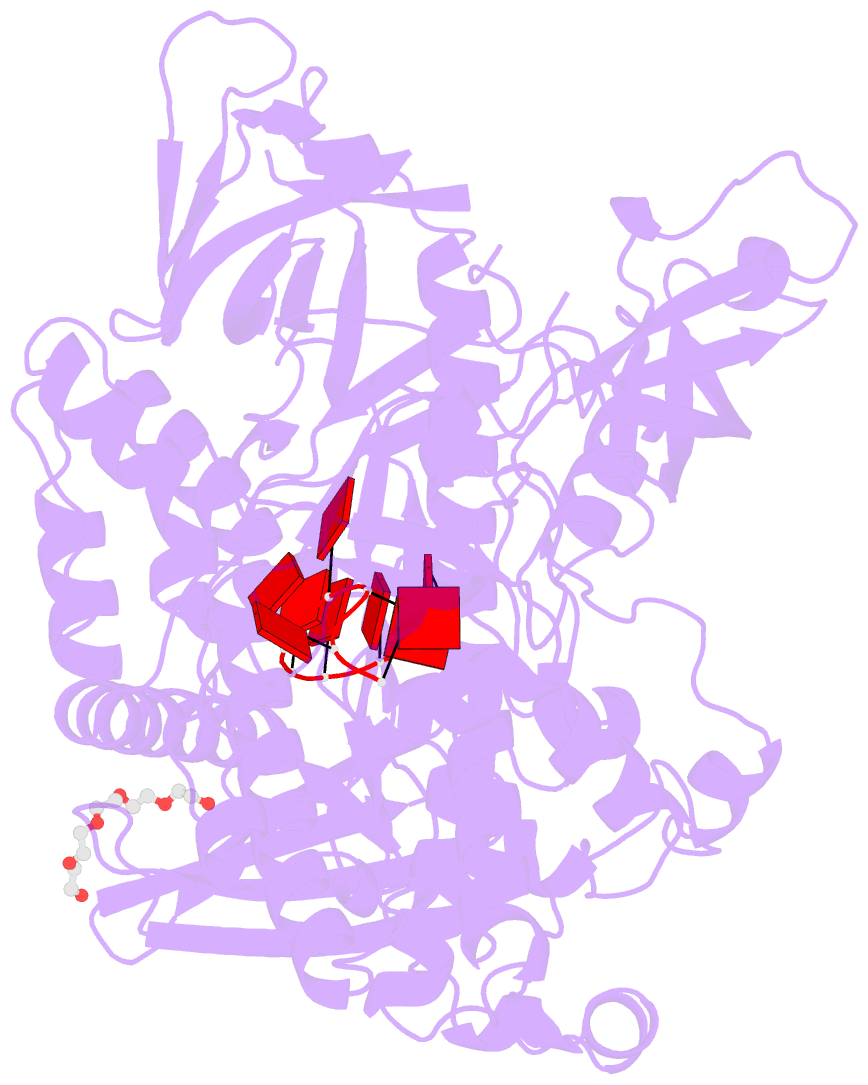
- Crystal structure of sc rrp44
- Reference
- Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E (2008): "Structure of the Active Subunit of the Yeast Exosome Core, Rrp44: Diverse Modes of Substrate Recruitment in the Rnase II Nuclease Family." Mol.Cell, 29, 717. doi: 10.1016/J.MOLCEL.2008.02.018.
- Abstract
- The eukaryotic exosome is a macromolecular complex essential for RNA processing and decay. It has recently been shown that the RNase activity of the yeast exosome core can be mapped to a single subunit, Rrp44, which processively degrades single-stranded RNAs as well as RNAs containing secondary structures. Here we present the 2.3 A resolution crystal structure of S. cerevisiae Rrp44 in complex with single-stranded RNA. Although Rrp44 has a linear domain organization similar to bacterial RNase II, in three dimensions the domains have a different arrangement. The three domains of the classical nucleic-acid-binding OB fold are positioned on the catalytic domain such that the RNA-binding path observed in RNase II is occluded. Instead, RNA is threaded to the catalytic site via an alternative route suggesting a mechanism for RNA-duplex unwinding. The structure provides a molecular rationale for the observed biochemical properties of the RNase R family of nucleases.