Summary information and primary citation
- PDB-id
- 2vs7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.05 Å)
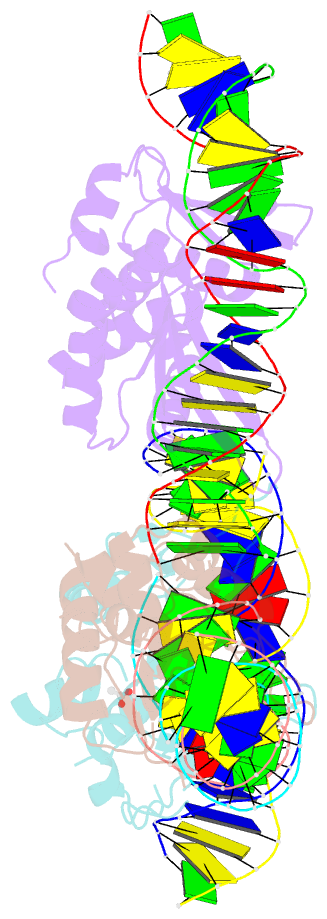
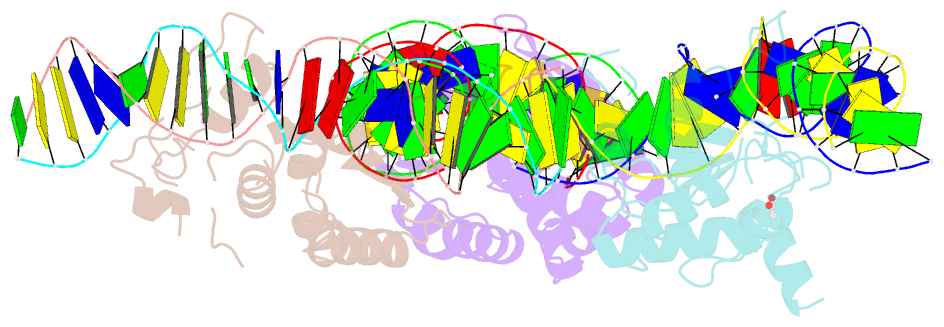
- Summary
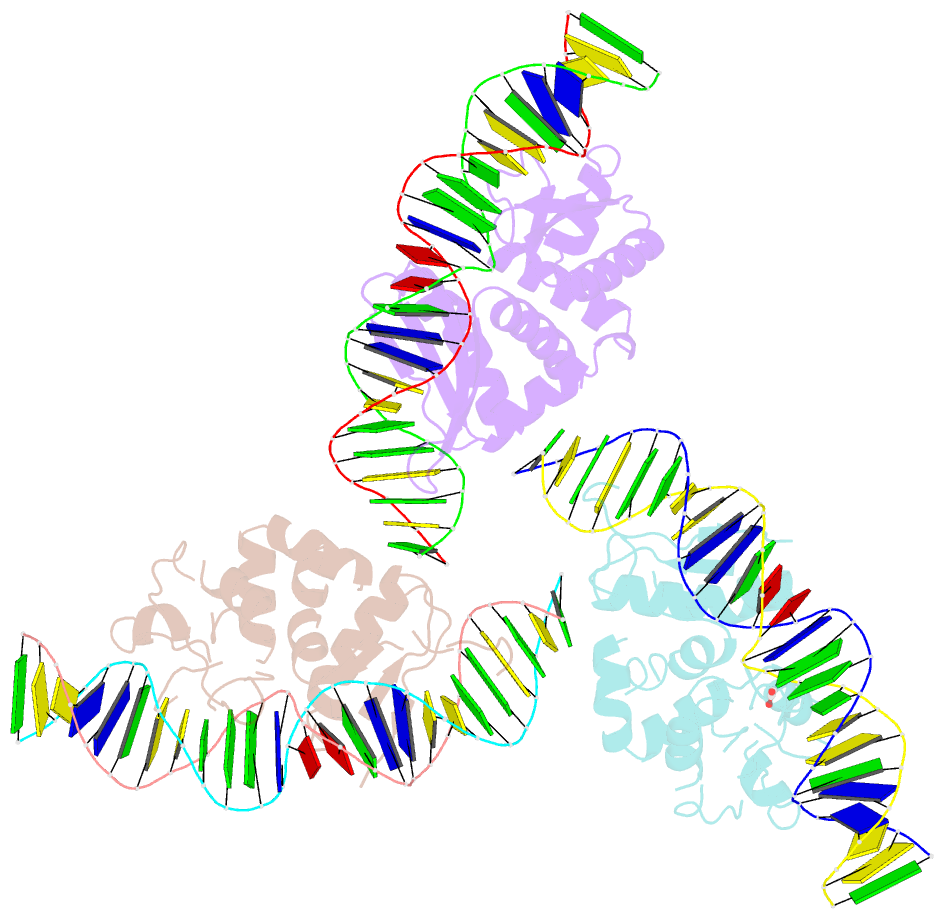
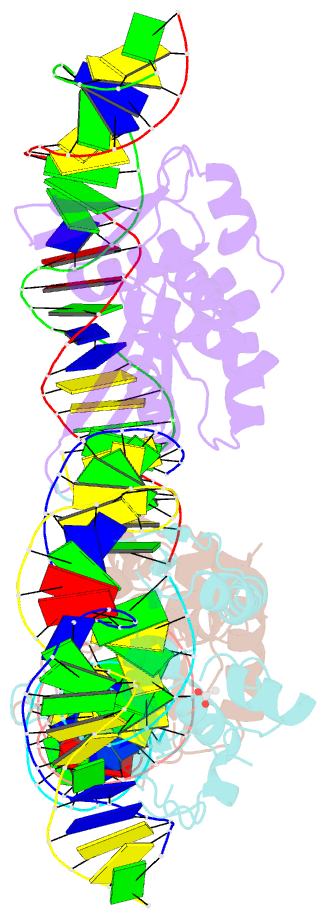
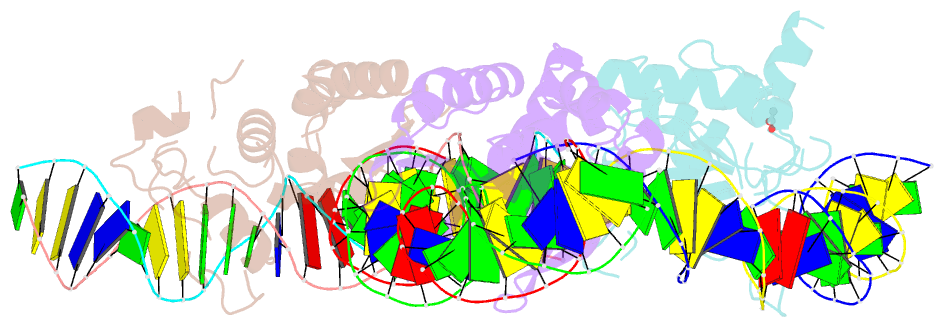
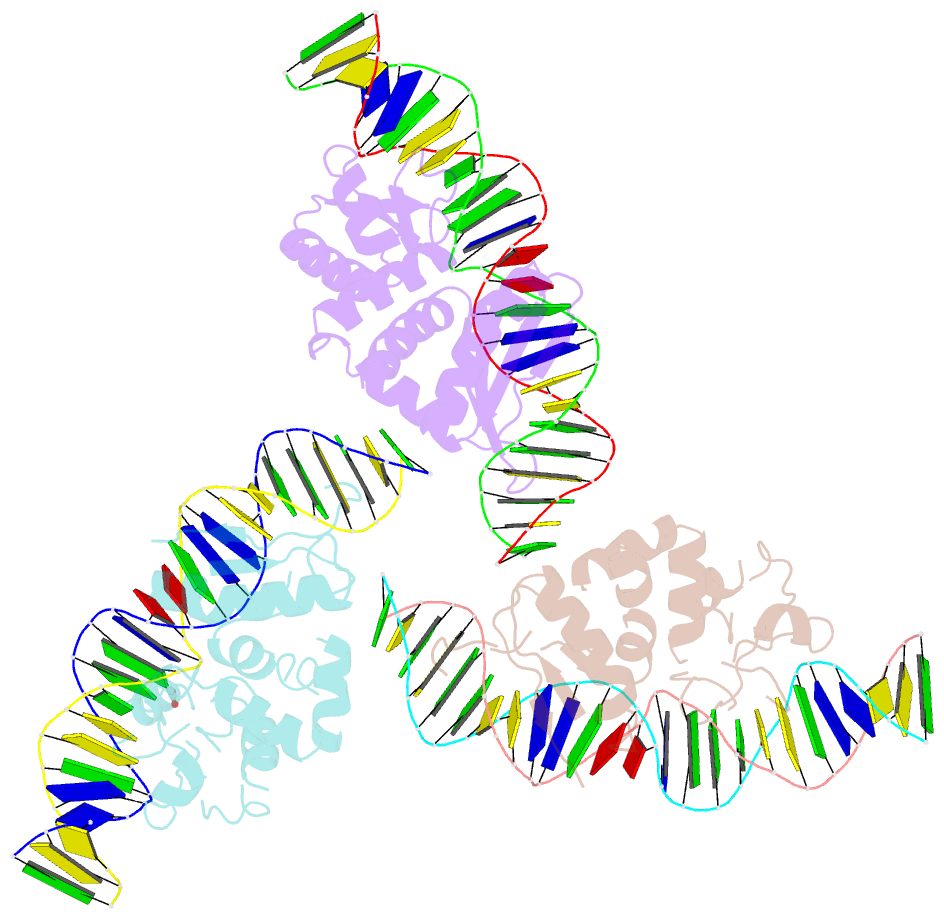
- The crystal structure of i-dmoi in complex with DNA and ca
- Reference
- Marcaida MJ, Prieto J, Redondo P, Nadra AD, Alibes A, Serrano L, Grizot S, Duchateau P, Paques F, Blanco FJ, Montoya G (2008): "Crystal Structure of I-Dmoi in Complex with its Target DNA Provides New Insights Into Meganuclease Engineering." Proc.Natl.Acad.Sci.USA, 105, 16888. doi: 10.1073/PNAS.0804795105.
- Abstract
- Homing endonucleases, also known as meganucleases, are sequence-specific enzymes with large DNA recognition sites. These enzymes can be used to induce efficient homologous gene targeting in cells and plants, opening perspectives for genome engineering with applications in a wide series of fields, ranging from biotechnology to gene therapy. Here, we report the crystal structures at 2.0 and 2.1 A resolution of the I-DmoI meganuclease in complex with its substrate DNA before and after cleavage, providing snapshots of the catalytic process. Our study suggests that I-DmoI requires only 2 cations instead of 3 for DNA cleavage. The structure sheds light onto the basis of DNA binding, indicating key residues responsible for nonpalindromic target DNA recognition. In silico and in vivo analysis of the I-DmoI DNA cleavage specificity suggests that despite the relatively few protein-base contacts, I-DmoI is highly specific when compared with other meganucleases. Our data open the door toward the generation of custom endonucleases for targeted genome engineering using the monomeric I-DmoI scaffold.