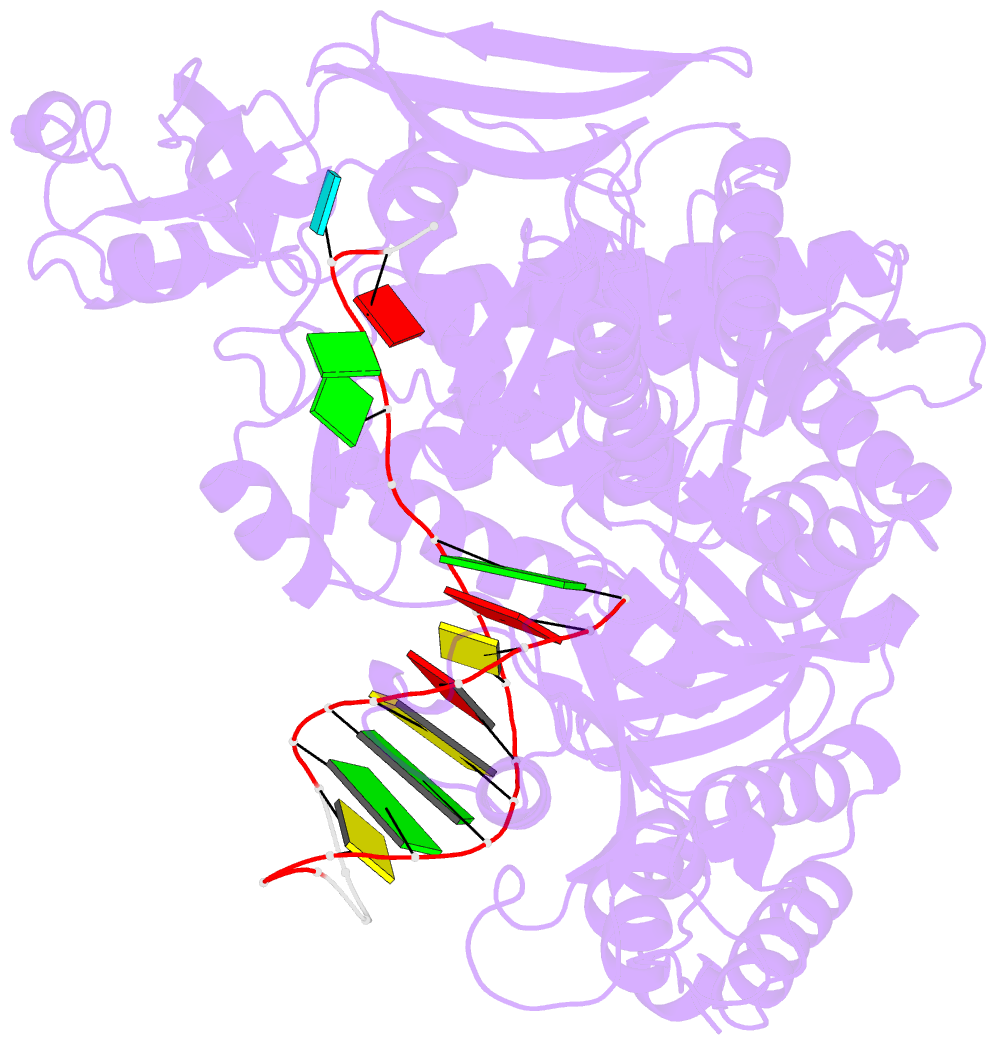
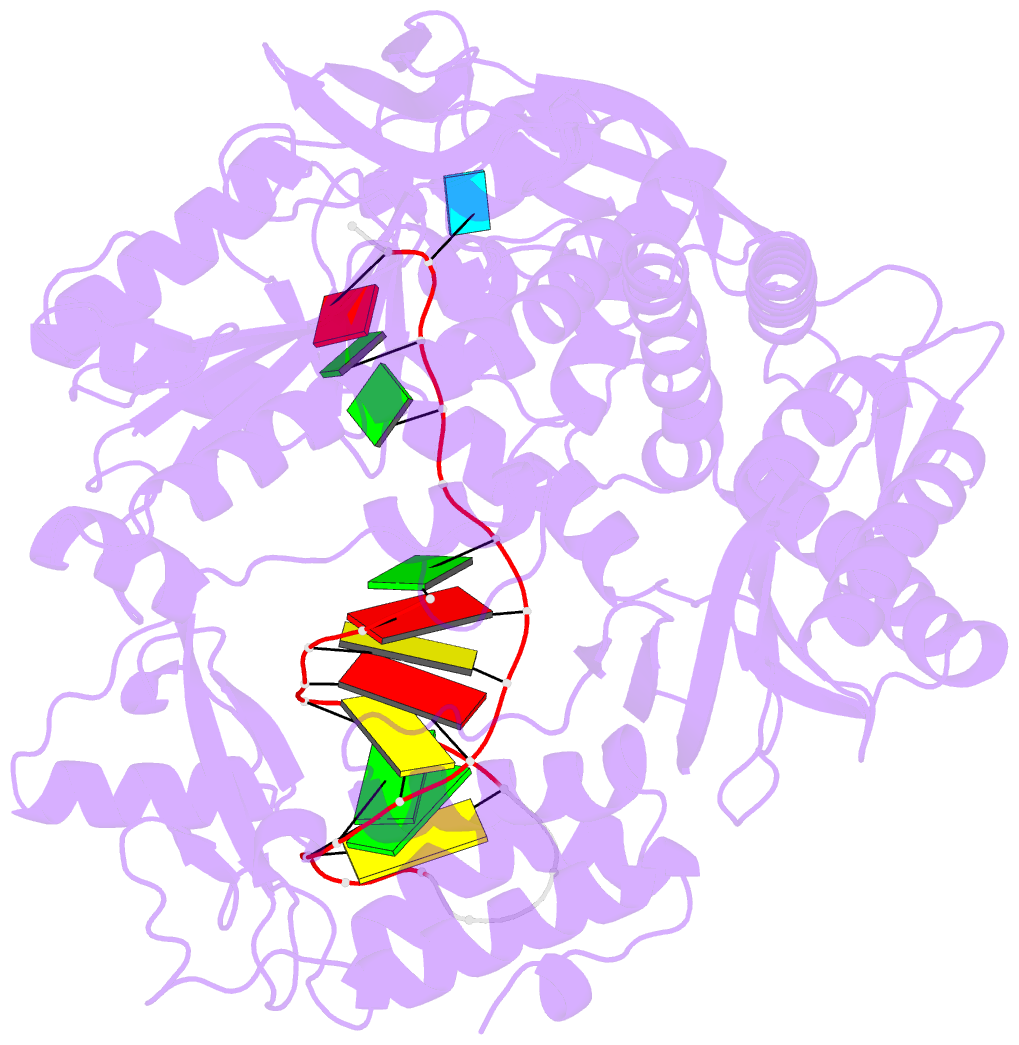
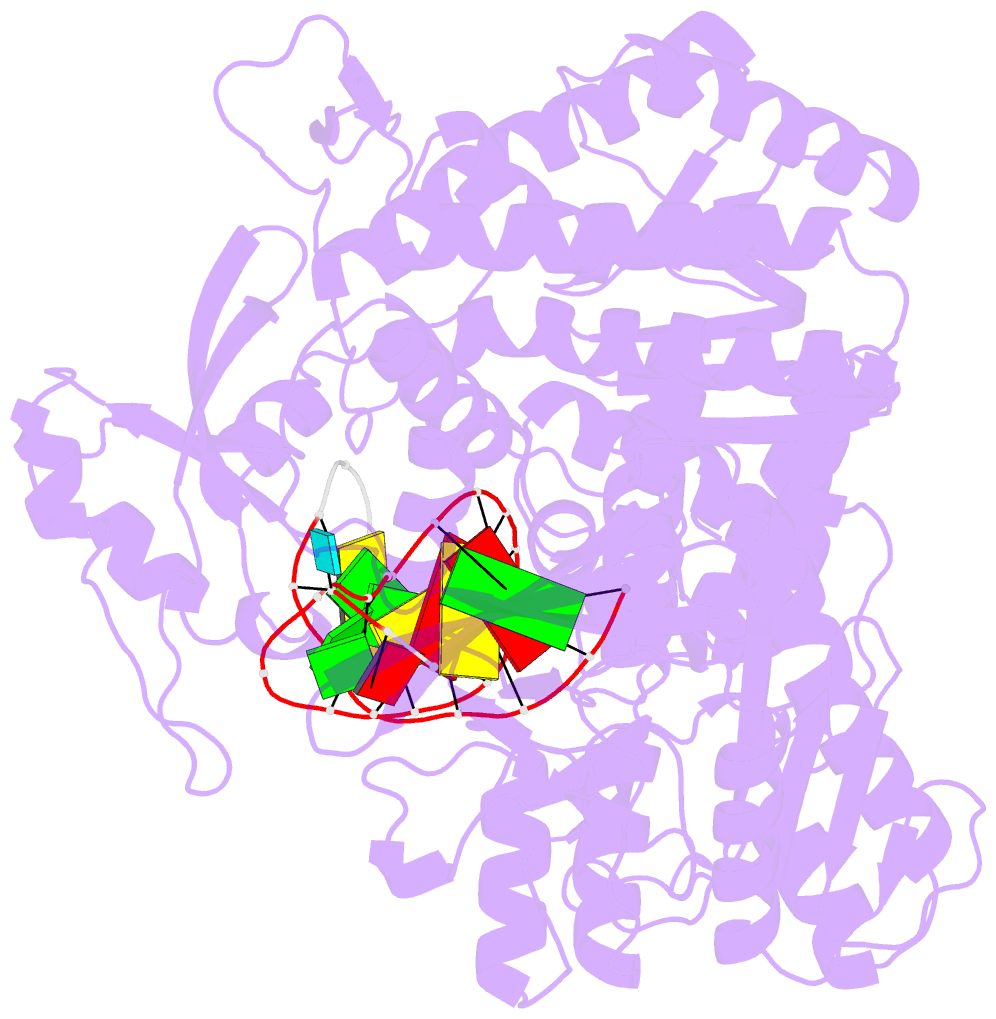
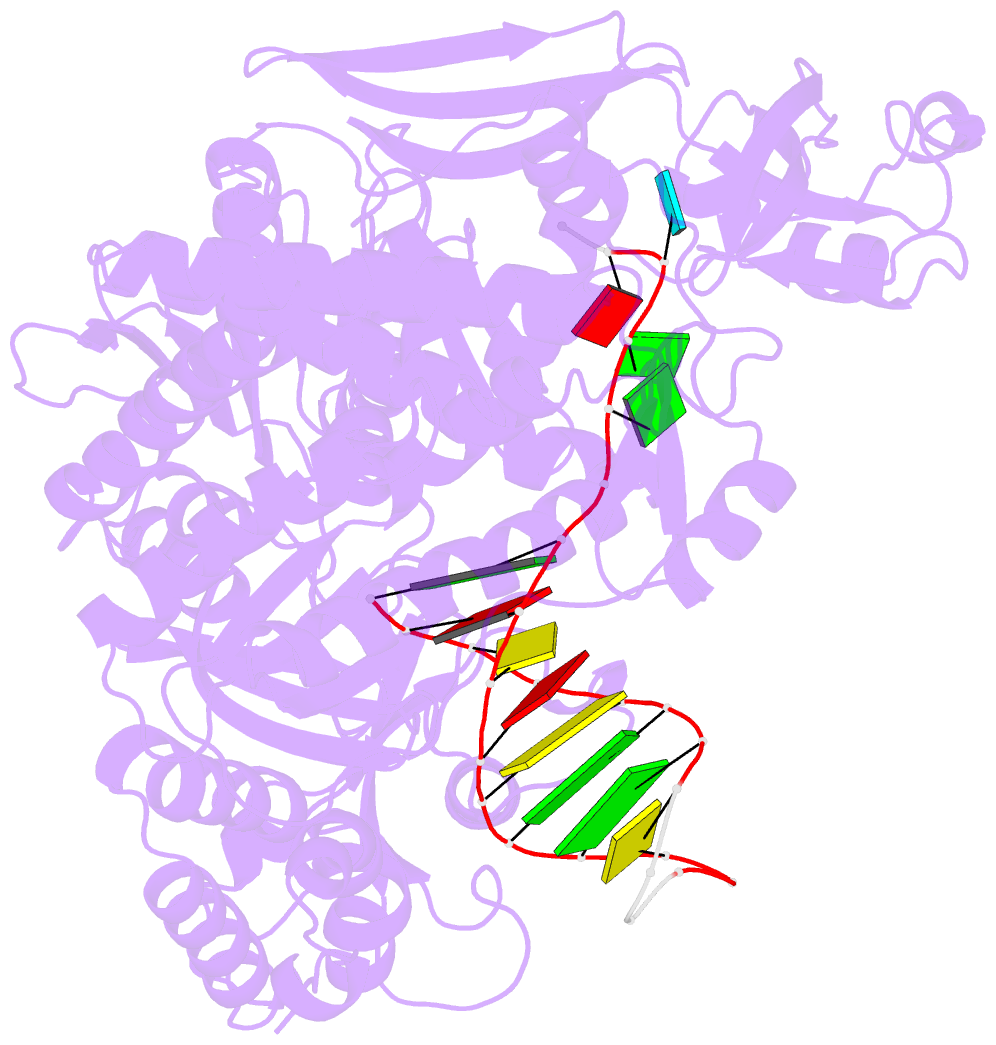
Summary information and primary citation
- PDB-id
- 2vwj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA replication
- Method
- X-ray (2.78 Å)
- Summary
- Uracil recognition in archaeal DNA polymerases captured by x-ray crystallography.
- Reference
- Firbank SJ, Wardle J, Heslop P, Lewis RJ, Connolly BA (2008): "Uracil Recognition in Archaeal DNA Polymerases Captured by X-Ray Crystallography." J.Mol.Biol., 381, 529. doi: 10.1016/J.JMB.2008.06.004.
- Abstract
- Archaeal family B DNA polymerases bind tightly to template-strand uracil and stall replication on encountering the pro-mutagenic base. This article describes an X-ray crystal structure, at 2.8 A resolution, of Thermococcus gorgonarius polymerase in complex with a DNA primer-template containing uracil in the single-stranded region. The DNA backbone is distorted to position the uracil deeply within a pocket, located in the amino-terminal domain of the polymerase. Specificity arises from a combination of hydrogen bonds between the protein backbone and uracil, with the pocket shaped to prevent the stable binding of the four standard DNA bases. Strong interactions are seen with the two phosphates that flank the uracil and the structure gives clues concerning the coupling of uracil binding to the halting of replication. The importance of key amino acids, identified by the analysis of the structure and their conservation between archaeal polymerases, was confirmed by site-directed mutagenesis. The crystal structure of V93Q, a polymerase variant that no longer recognises uracil, is also reported, explaining the V93Q phenotype by the steric exclusion of uracil from the pocket.