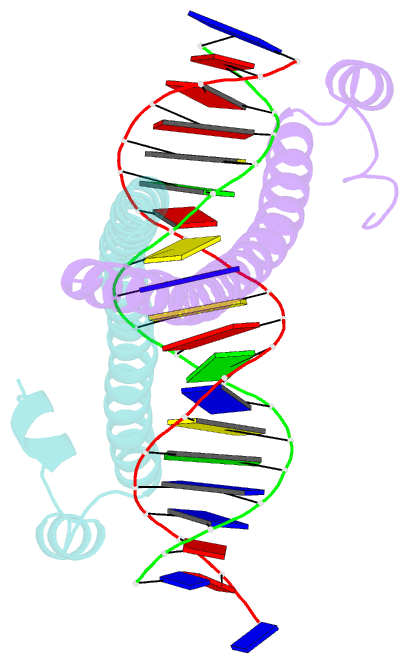
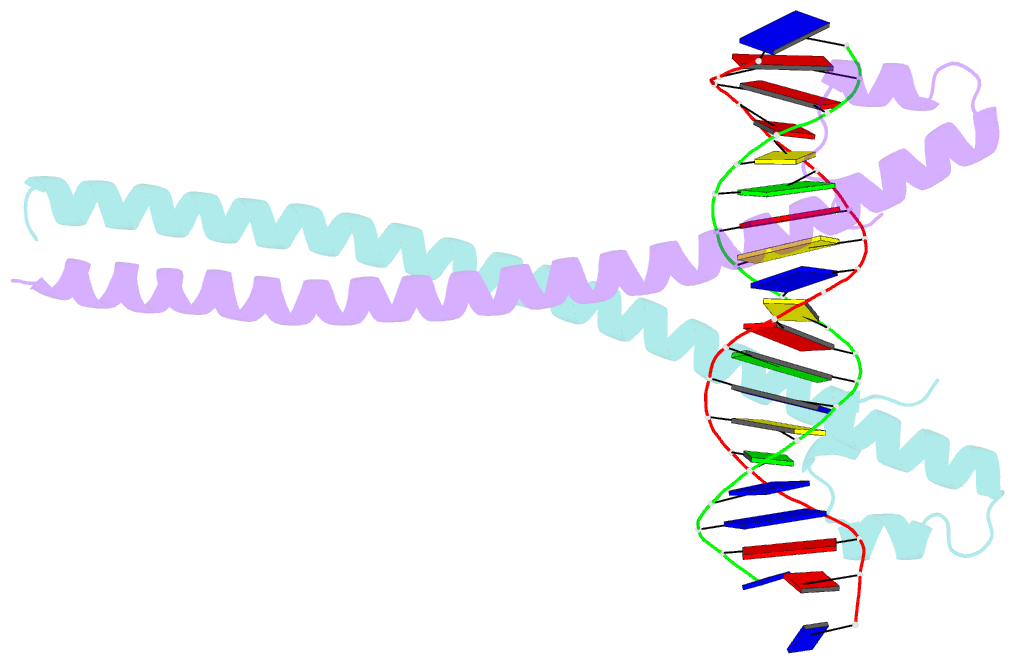
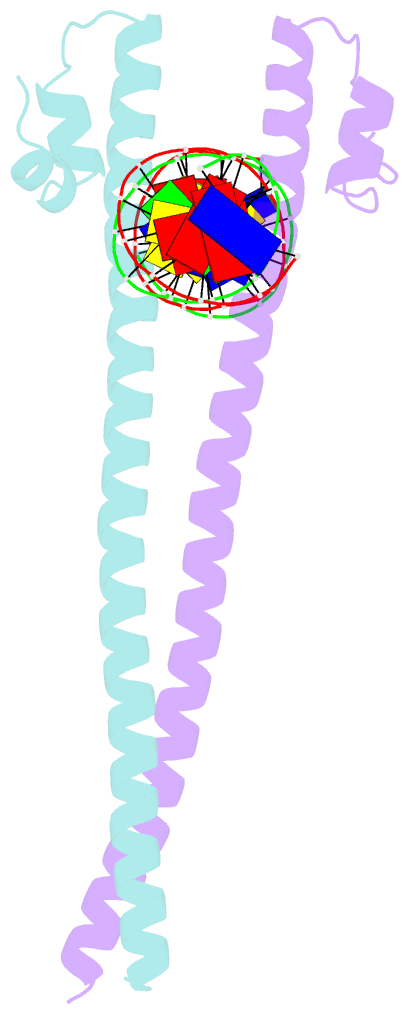
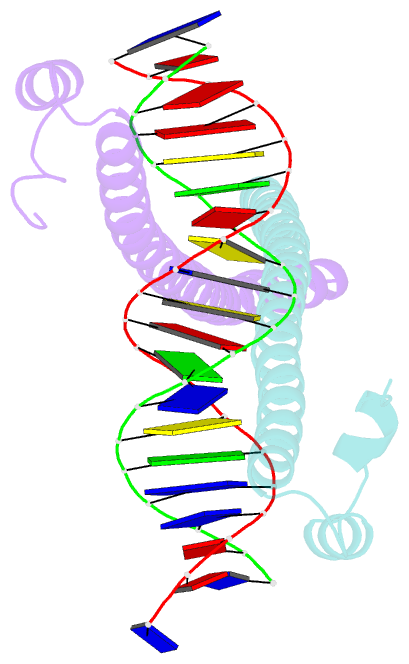
Summary information and primary citation
- PDB-id
- 2wty; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (2.9 Å)
- Summary
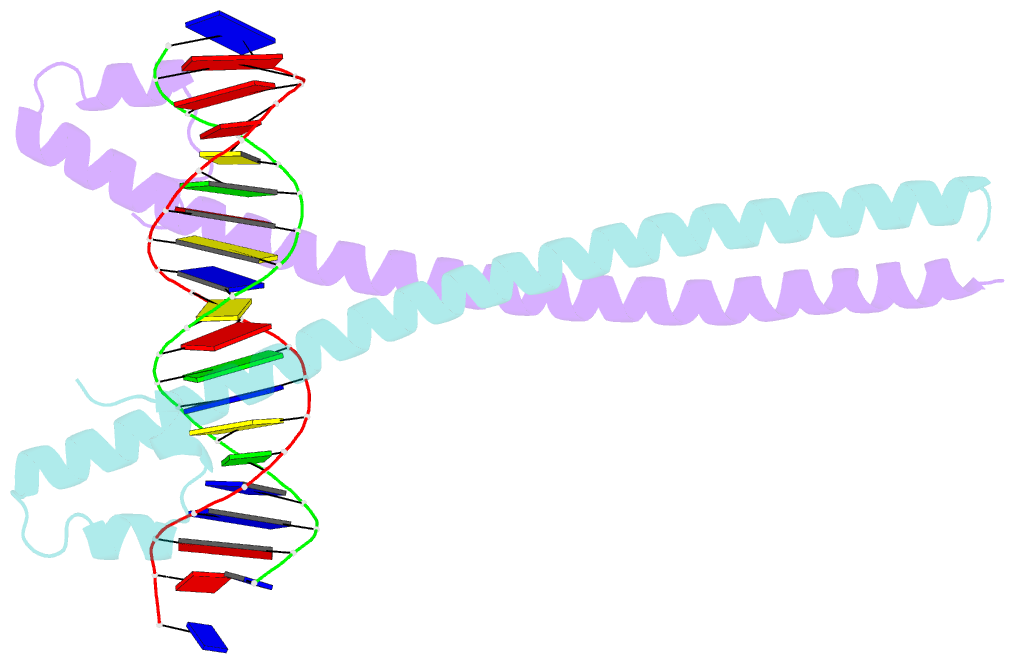
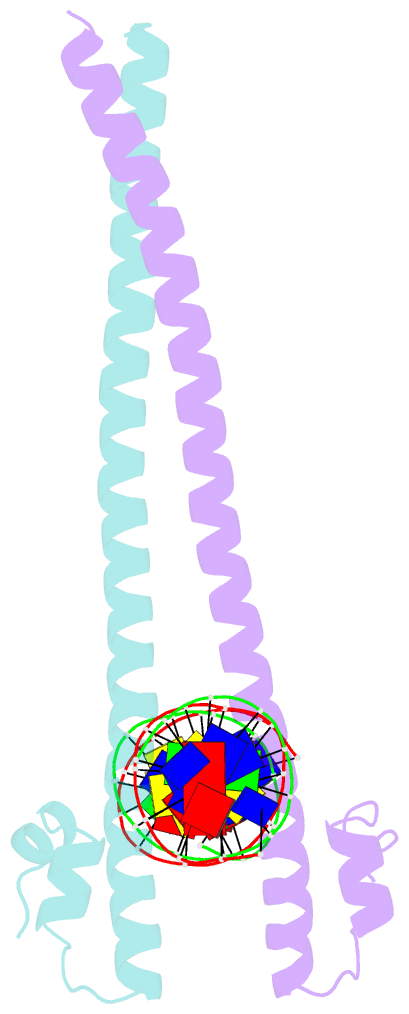
- Crystal structure of the homodimeric mafb in complex with the t-mare binding site
- Reference
- Pogenberg V, Consani Textor L, Vanhille L, Holton SJ, Sieweke MH, Wilmanns M (2014): "Design of a bZIP Transcription Factor with Homo/Heterodimer-Induced DNA-Binding Preference." Structure, 22, 466. doi: 10.1016/J.STR.2013.12.017.
- Abstract
- The ability of basic leucine zipper transcription factors for homo- or heterodimerization provides a paradigm for combinatorial control of eukaryotic gene expression. It has been unclear, however, how facultative dimerization results in alternative DNA-binding repertoires on distinct regulatory elements. To unravel the molecular basis of such coupled preferences, we determined two high-resolution structures of the transcription factor MafB as a homodimer and as a heterodimer with c-Fos bound to variants of the Maf-recognition element. The structures revealed several unexpected and dimer-specific coiled-coil-heptad interactions. Based on these findings, we have engineered two MafB mutants with opposite dimerization preferences. One of them showed a strong preference for MafB/c-Fos heterodimerization and enabled selection of heterodimer-favoring over homodimer-specific Maf-recognition element variants. Our data provide a concept for transcription factor design to selectively activate dimer-specific pathways and binding repertoires.