Summary information and primary citation
- PDB-id
- 2x1f; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-RNA
- Method
- X-ray (1.6 Å)
- Summary
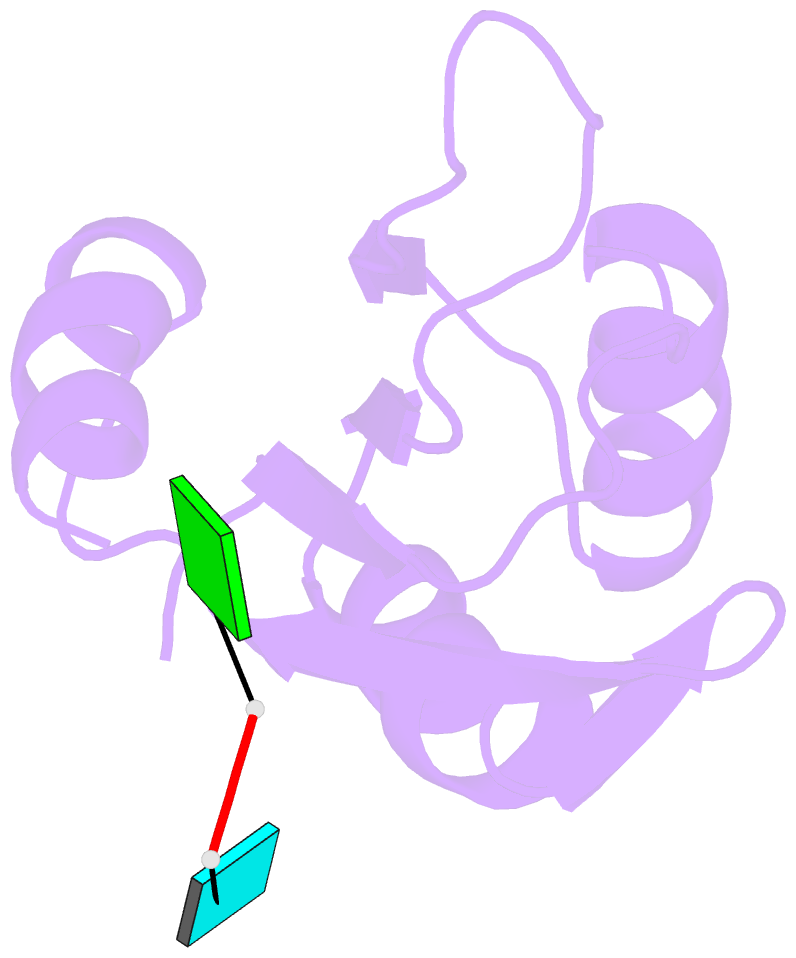
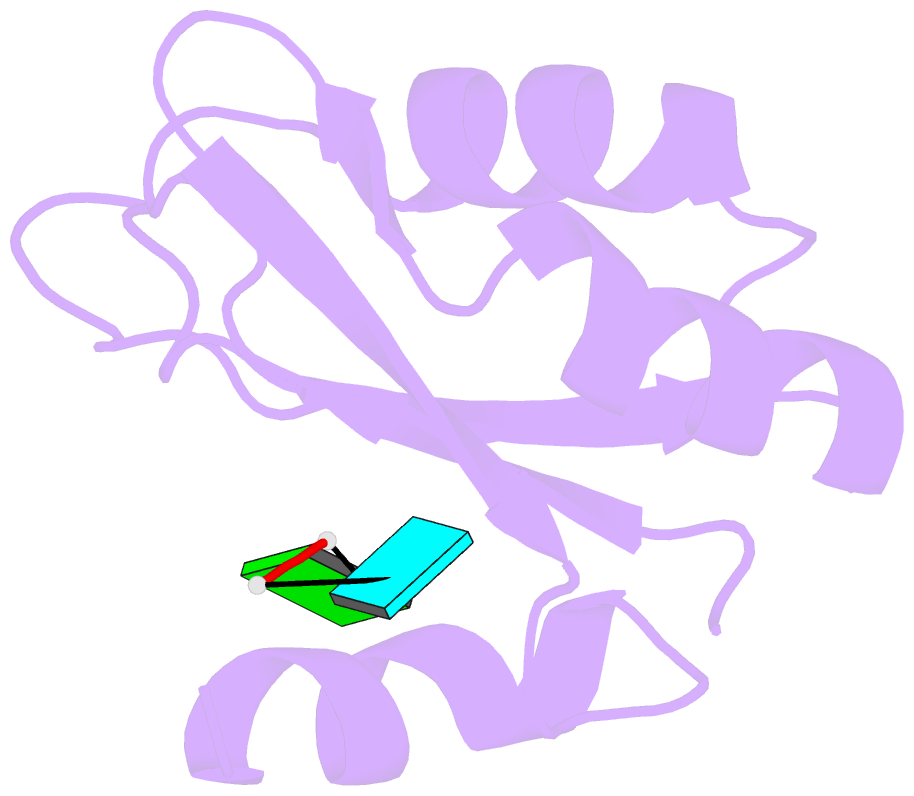
- Structure of rna15 rrm with bound RNA (gu)
- Reference
- Pancevac C, Goldstone DC, Ramos A, Taylor IA (2010): "Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors." Nucleic Acids Res., 38, 3119. doi: 10.1093/NAR/GKQ002.
- Abstract
- Rna15 is a core subunit of cleavage factor IA (CFIA), an essential transcriptional 3'-end processing factor from Saccharomyces cerevisiae. CFIA is required for polyA site selection/cleavage targeting RNA sequences that surround polyadenylation sites in the 3'-UTR of RNA polymerase-II transcripts. RNA recognition by CFIA is mediated by an RNA recognition motif (RRM) contained in the Rna15 subunit of the complex. We show here that Rna15 has a strong and unexpected preference for GU containing RNAs and reveal the molecular basis for a base selectivity mechanism that accommodates G or U but discriminates against C and A bases. This mode of base selectivity is rather different to that observed in other RRM-RNA structures and is structurally conserved in CstF64, the mammalian counterpart of Rna15. Our observations provide evidence for a highly conserved mechanism of base recognition amongst the 3'-end processing complexes that interact with the U-rich or U/G-rich elements at 3'-end cleavage/polyadenylation sites.