Summary information and primary citation
- PDB-id
- 2xb2; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (3.4 Å)
- Summary
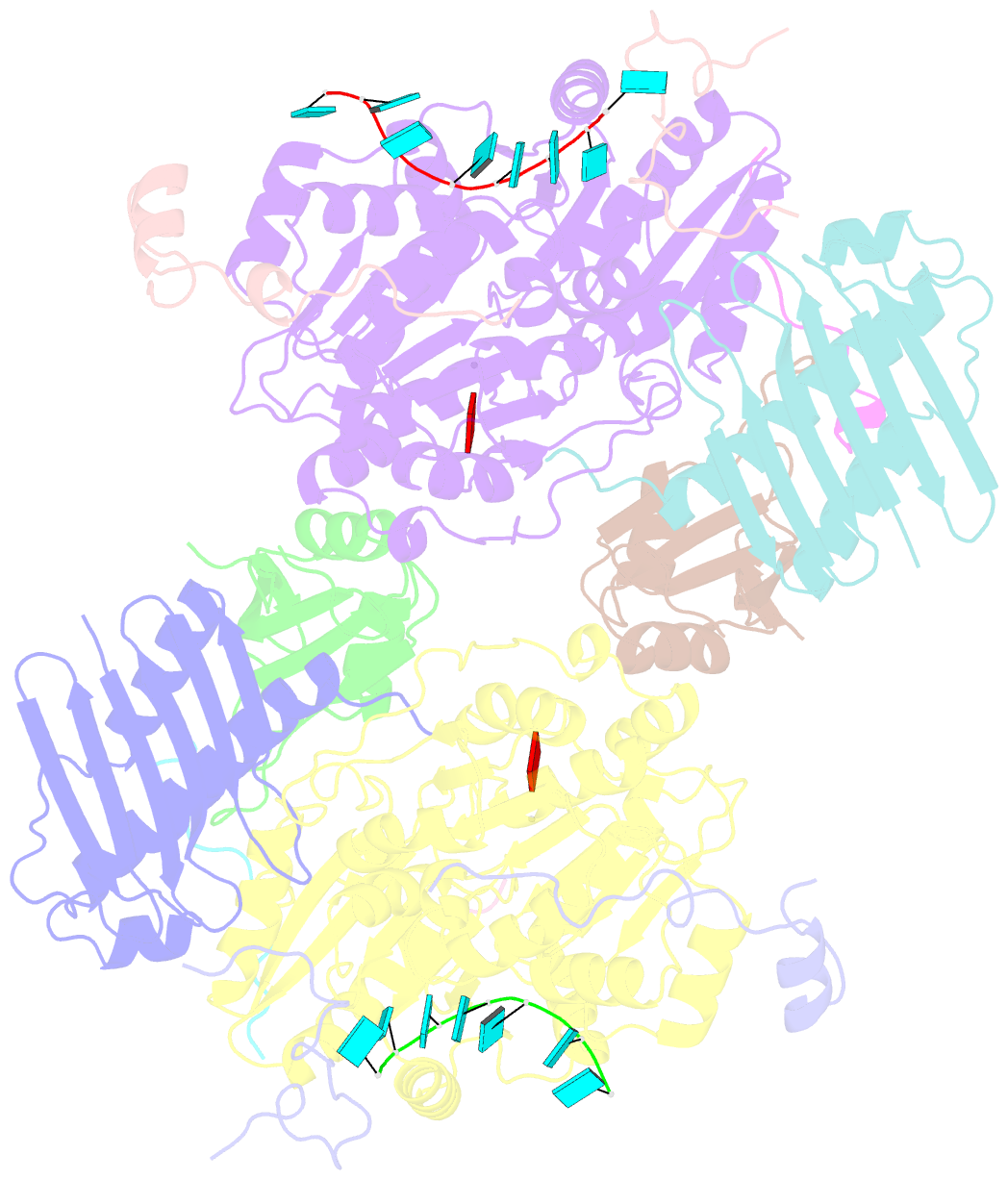
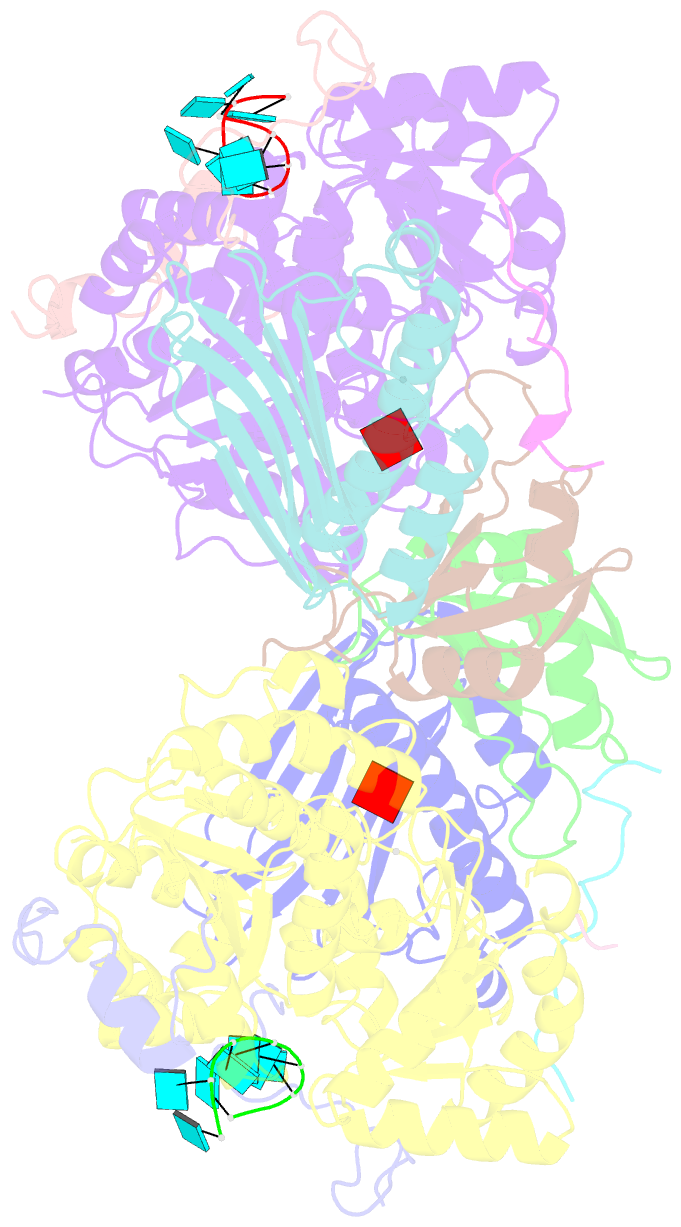
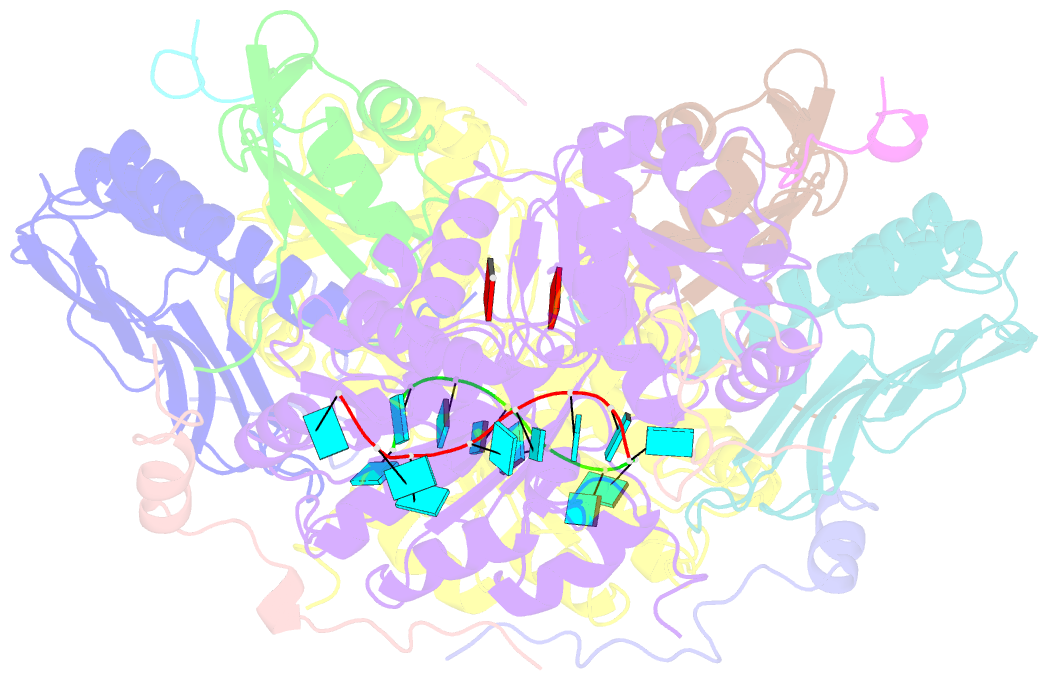
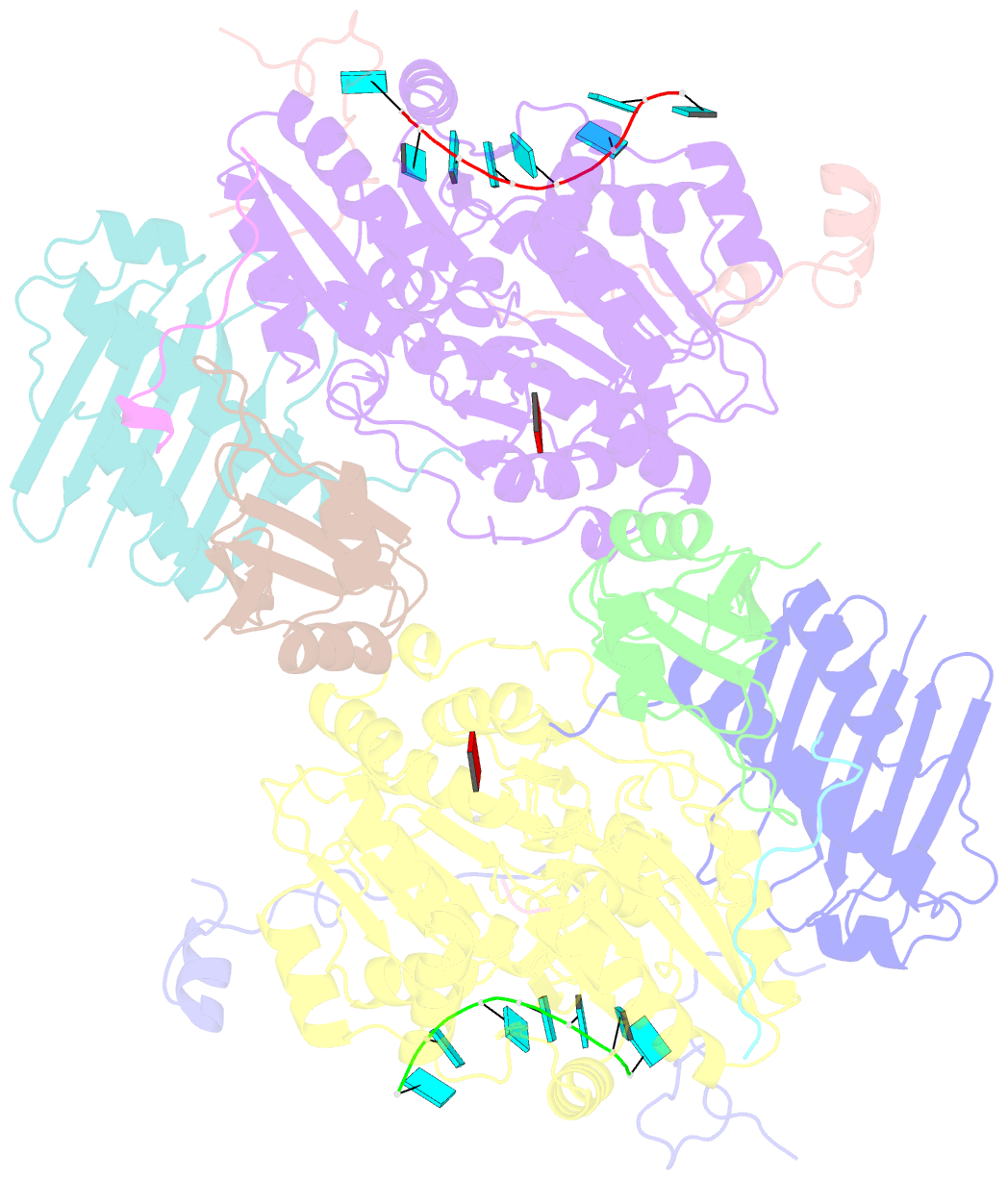
- Crystal structure of the core mago-y14-eif4aiii-barentsz-upf3b assembly shows how the ejc is bridged to the nmd machinery
- Reference
- Buchwald G, Ebert J, Basquin C, Sauliere J, Jayachandran U, Bono F, Le Hir H, Conti E (2010): "Insights Into the Recruitment of the Nmd Machinery from the Crystal Structure of a Core Ejc-Upf3B Complex." Proc.Natl.Acad.Sci.USA, 107, 10050. doi: 10.1073/PNAS.1000993107.
- Abstract
- In mammals, Up-frameshift proteins (UPFs) form a surveillance complex that interacts with the exon junction complex (EJC) to elicit nonsense-mediated mRNA decay (NMD). UPF3b is the component of the surveillance complex that bridges the interaction with the EJC. Here, we report the 3.4 A resolution crystal structure of a minimal UPF3b-EJC assembly, consisting of the interacting domains of five proteins (UPF3b, MAGO, Y14, eIF4AIII, and Barentsz) together with RNA and adenylyl-imidodiphosphate. Human UPF3b binds with the C-terminal domain stretched over a composite surface formed by eIF4AIII, MAGO, and Y14. Residues that affect NMD when mutated are found at the core interacting surfaces, whereas differences between UPF3b and UPF3a map at peripheral interacting residues. Comparison with the binding mode of the protein PYM underscores how a common molecular surface of MAGO and Y14 recognizes different proteins acting at different times in the same pathway. The binding mode to eIF4AIII identifies a surface hot spot that is used by different DEAD-box proteins to recruit their regulators.