Summary information and primary citation
- PDB-id
- 2xhb; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.72 Å)
- Summary
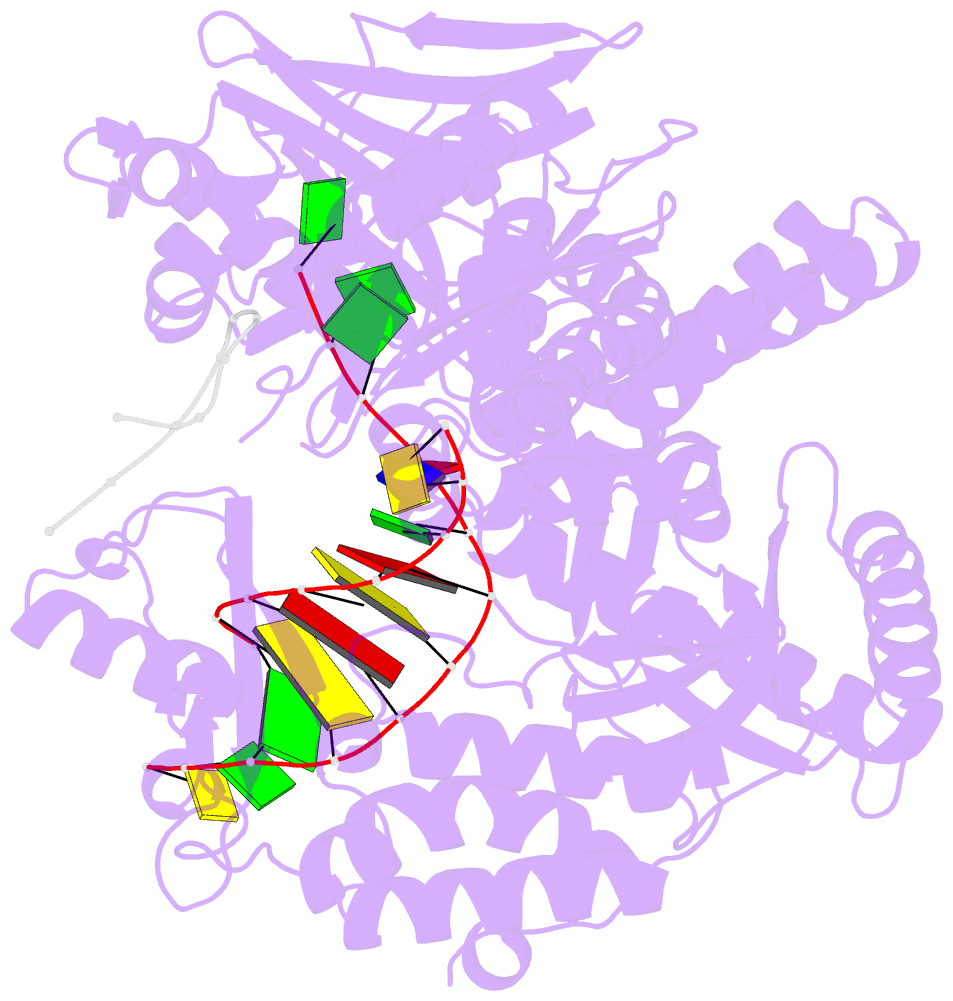
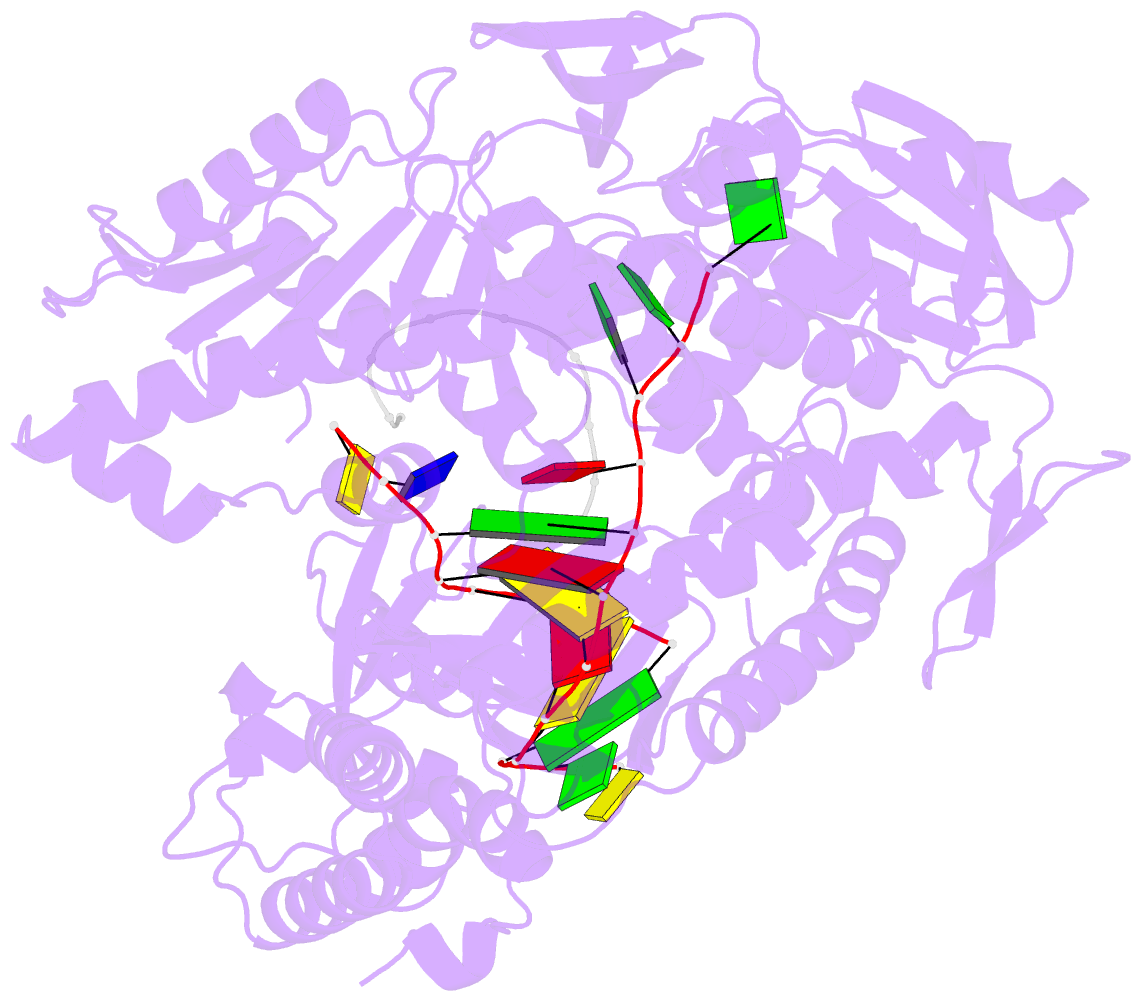
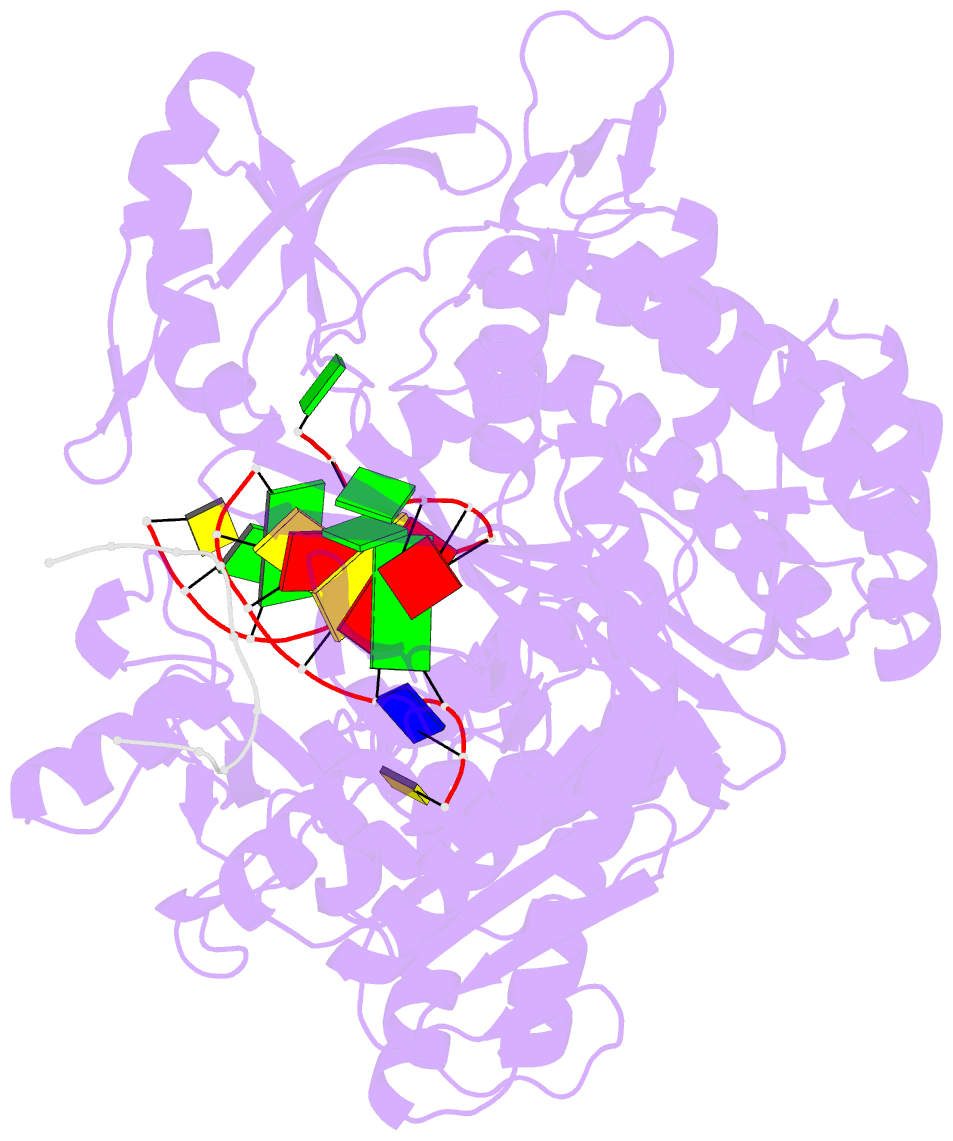
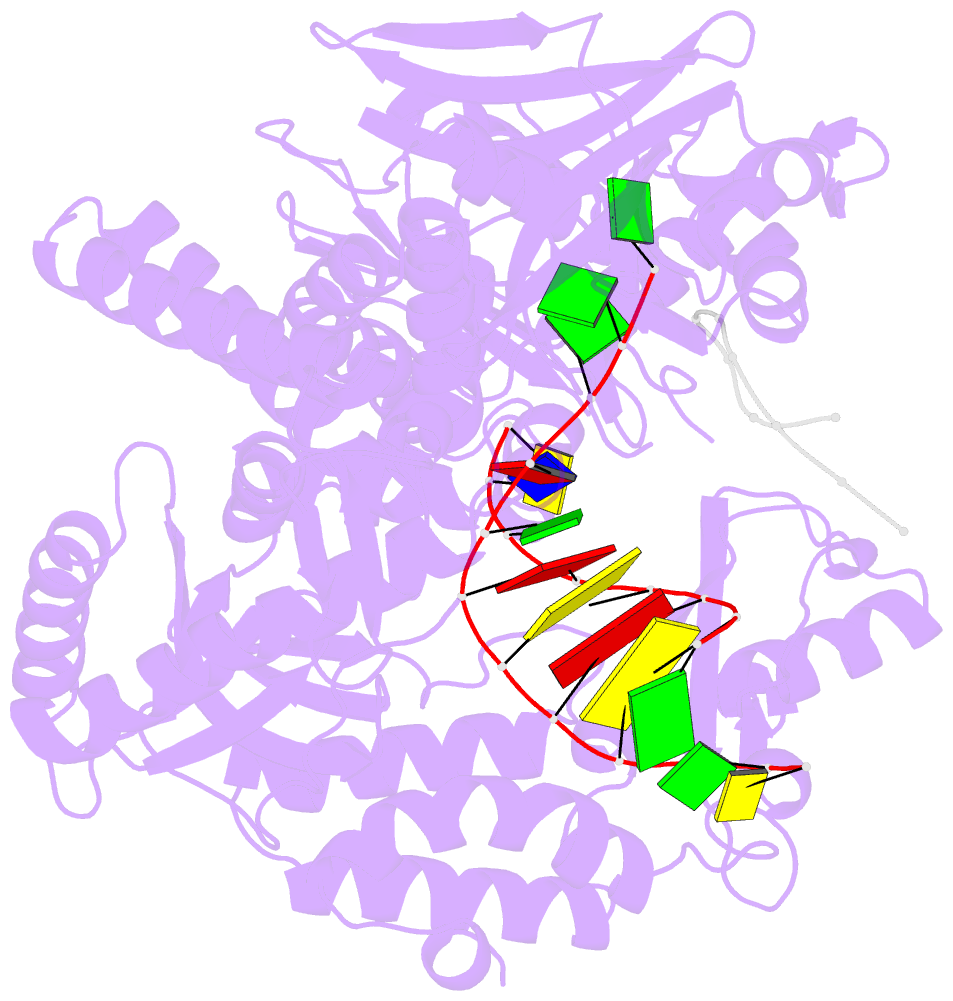
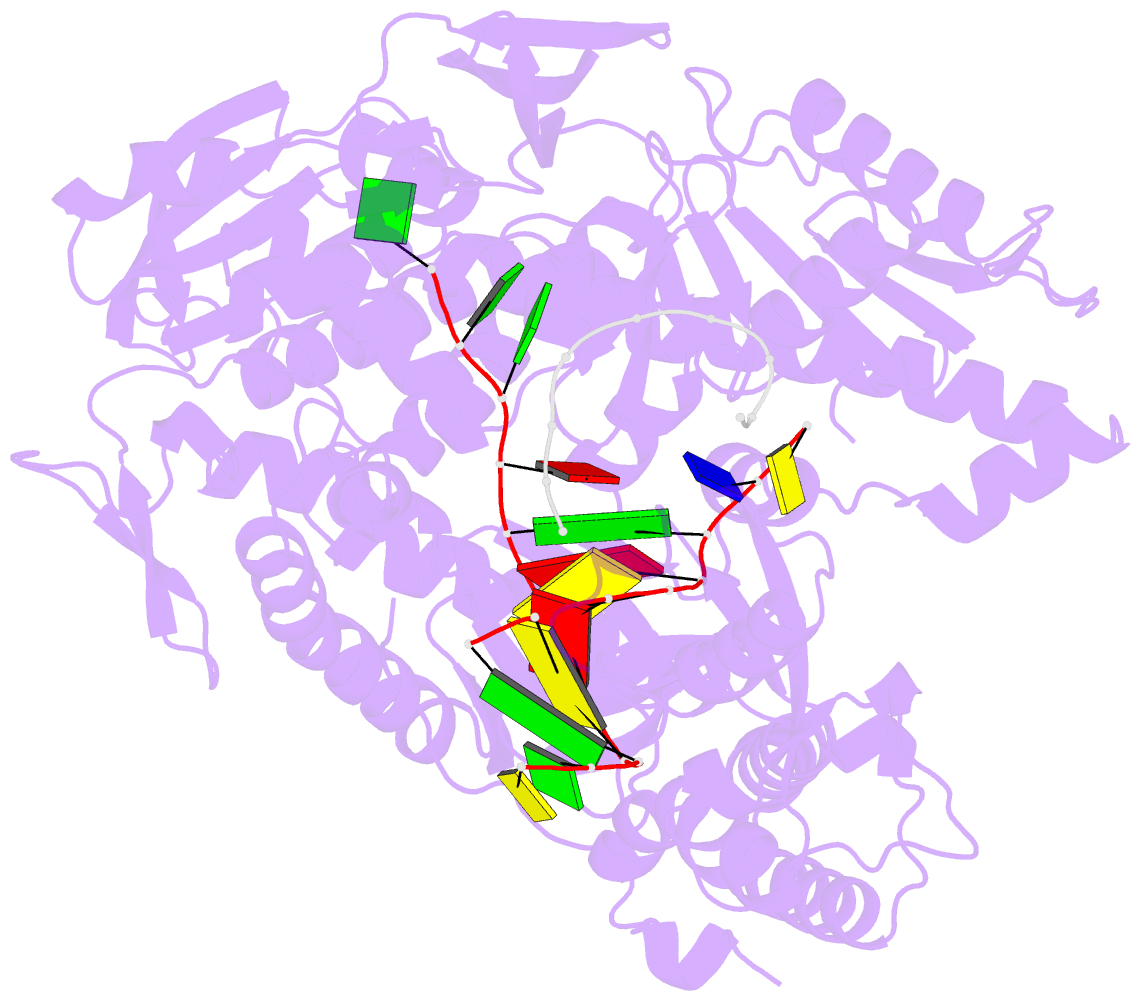
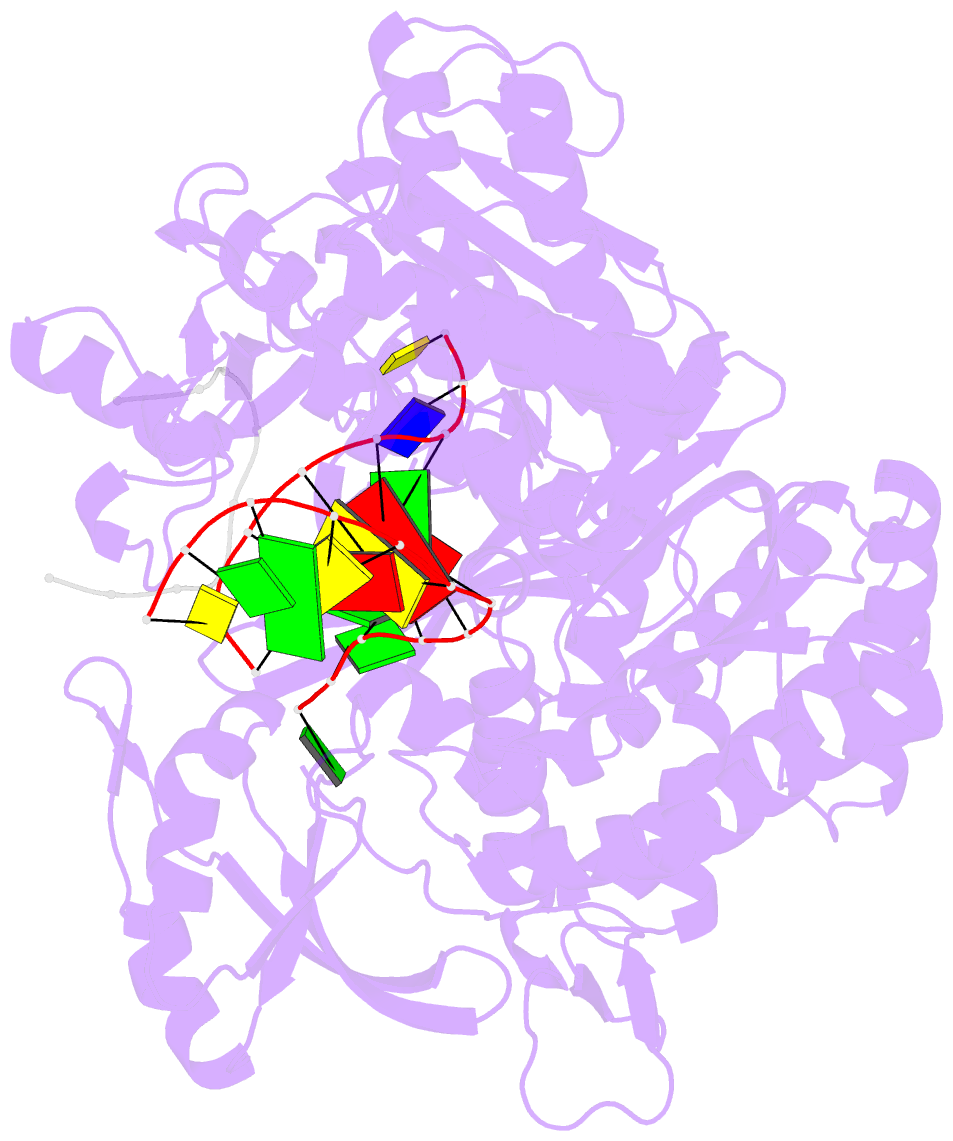
- Crystal structure of DNA polymerase from thermococcus gorgonarius in complex with hypoxanthine-containing DNA
- Reference
- Killelea T, Ghosh S, Tan SS, Heslop P, Firbank SJ, Kool ET, Connolly BA (2010): "Probing the Interaction of Archaeal DNA Polymerases with Deaminated Bases Using X-Ray Crystallography and Non-Hydrogen Bonding Isosteric Base Analogues." Biochemistry, 49, 5772. doi: 10.1021/BI100421R.
- Abstract
- Archaeal family-B DNA polymerases stall replication on encountering the pro-mutagenic bases uracil and hypoxanthine. This publication describes an X-ray crystal structure of Thermococcus gorgonarius polymerase in complex with a DNA containing hypoxanthine in the single-stranded region of the template, two bases ahead of the primer-template junction. Full details of the specific recognition of hypoxanthine are revealed, allowing a comparison with published data that describe uracil binding. The two bases are recognized by the same pocket, in the N-terminal domain, and make very similar protein-DNA interactions. Specificity for hypoxanthine (and uracil) arises from a combination of polymerase-base hydrogen bonds and shape fit between the deaminated bases and the pocket. The structure with hypoxanthine at position 2 explains the stimulation of the polymerase 3'-5' proofreading exonuclease, observed with deaminated bases at this location. A beta-hairpin element, involved in partitioning the primer strand between the polymerase and exonuclease active sites, inserts between the two template bases at the extreme end of the double-stranded DNA. This denatures the two complementary primer bases and directs the resulting 3' single-stranded extension toward the exonuclease active site. Finally, the relative importance of hydrogen bonding and shape fit in determining selectivity for deaminated bases has been examined using nonpolar isosteres. Affinity for both 2,4-difluorobenzene and fluorobenzimidazole, non-hydrogen bonding shape mimics of uracil and hypoxanthine, respectively, is strongly diminished, suggesting polar protein-base contacts are important. However, residual interaction with 2,4-difluorobenzene is seen, confirming a role for shape recognition.