Summary information and primary citation
- PDB-id
- 2xnr; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (1.6 Å)
- Summary
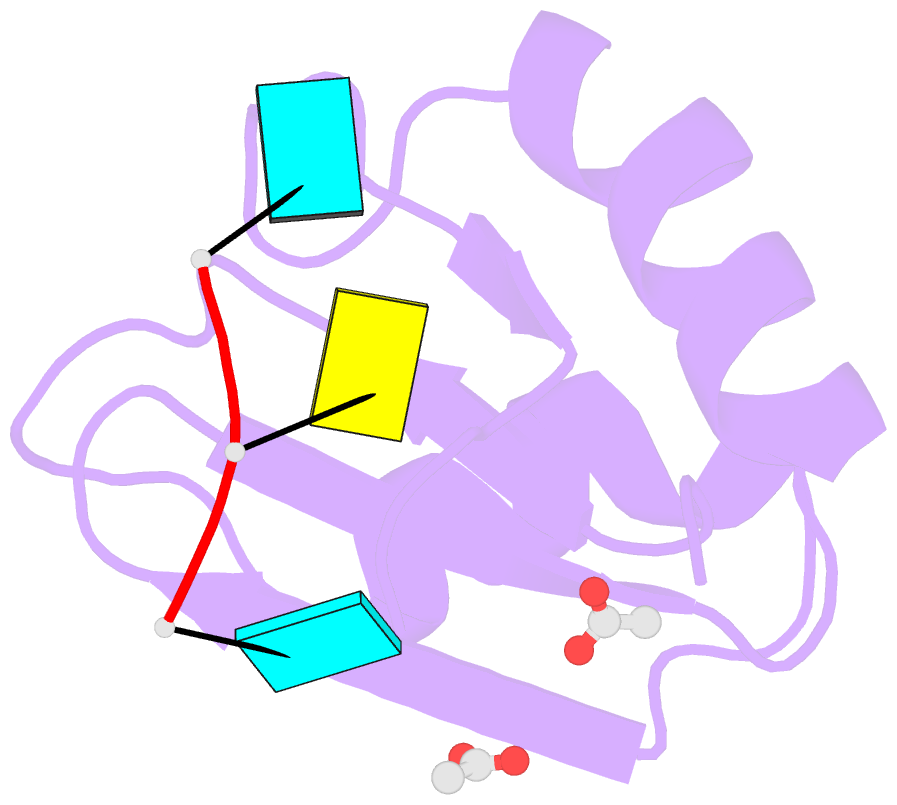
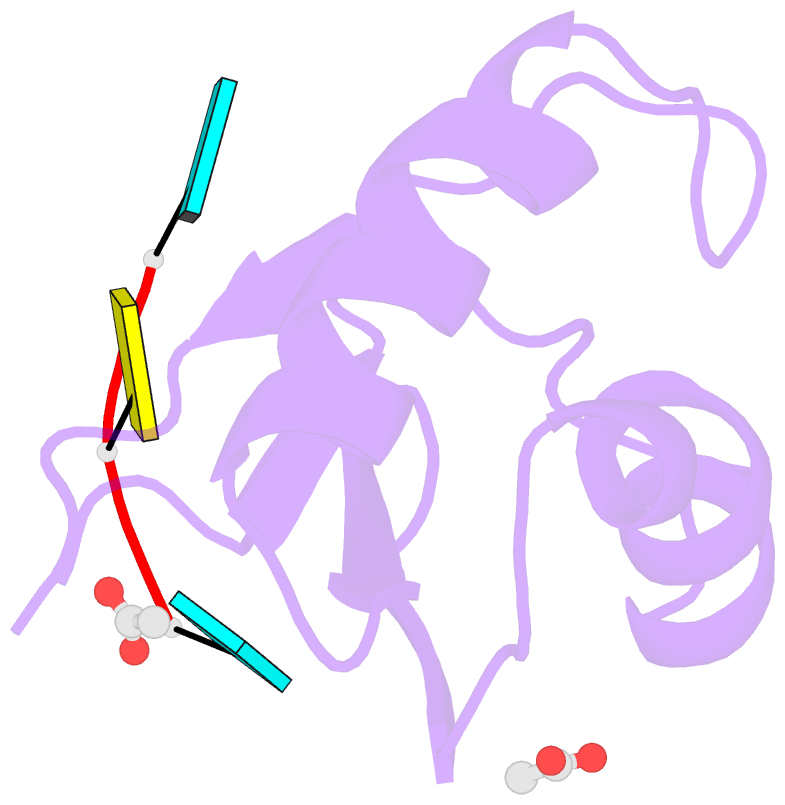
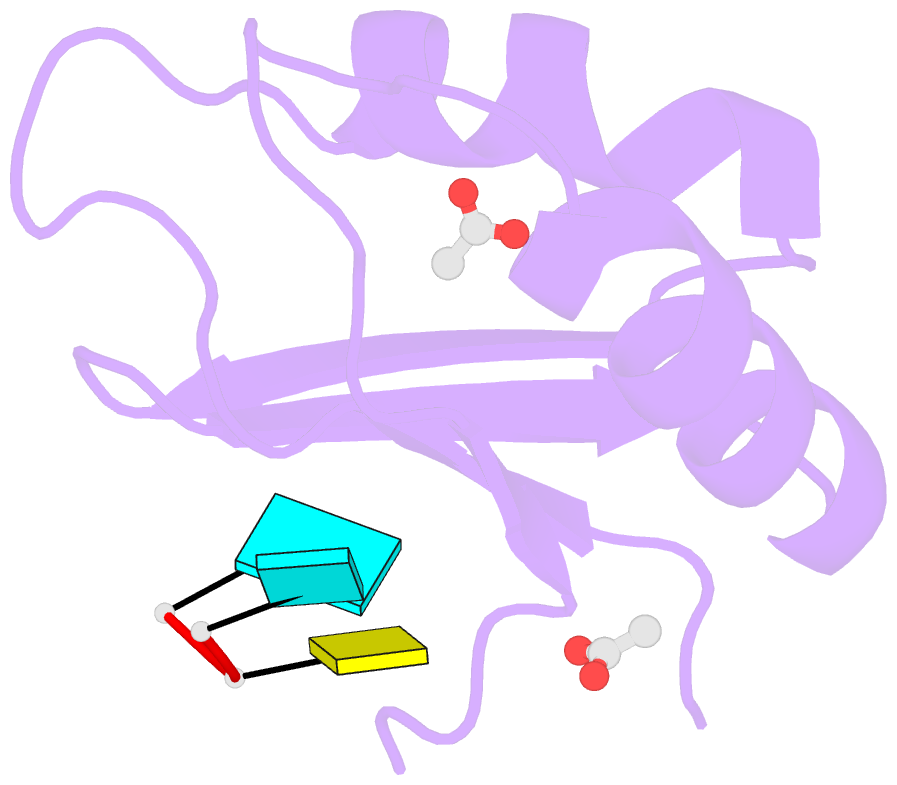
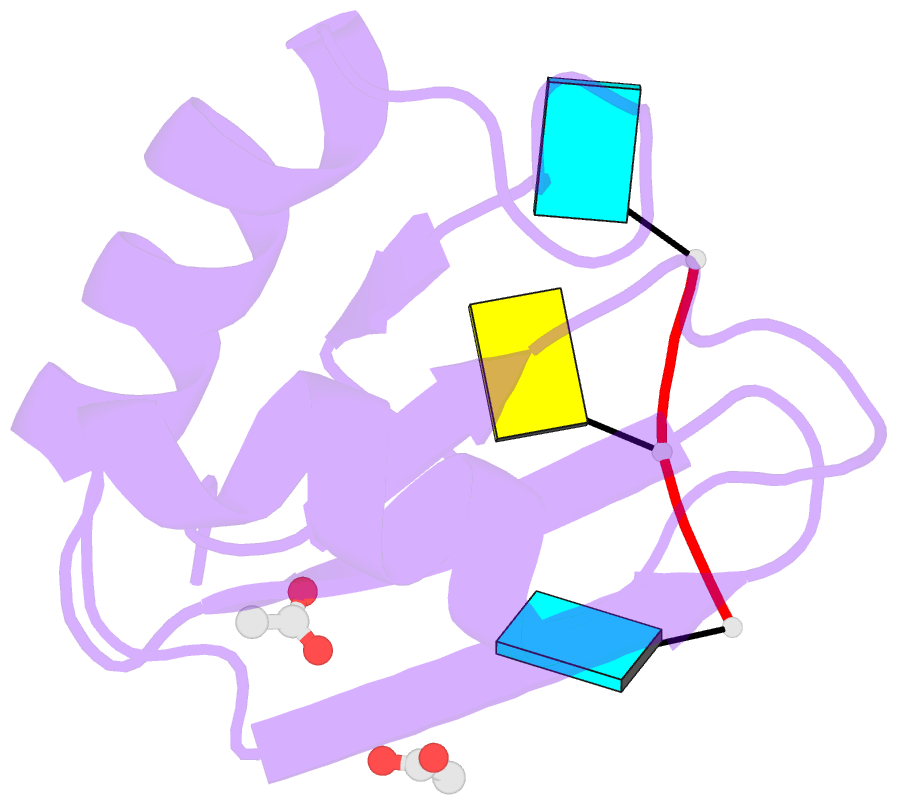
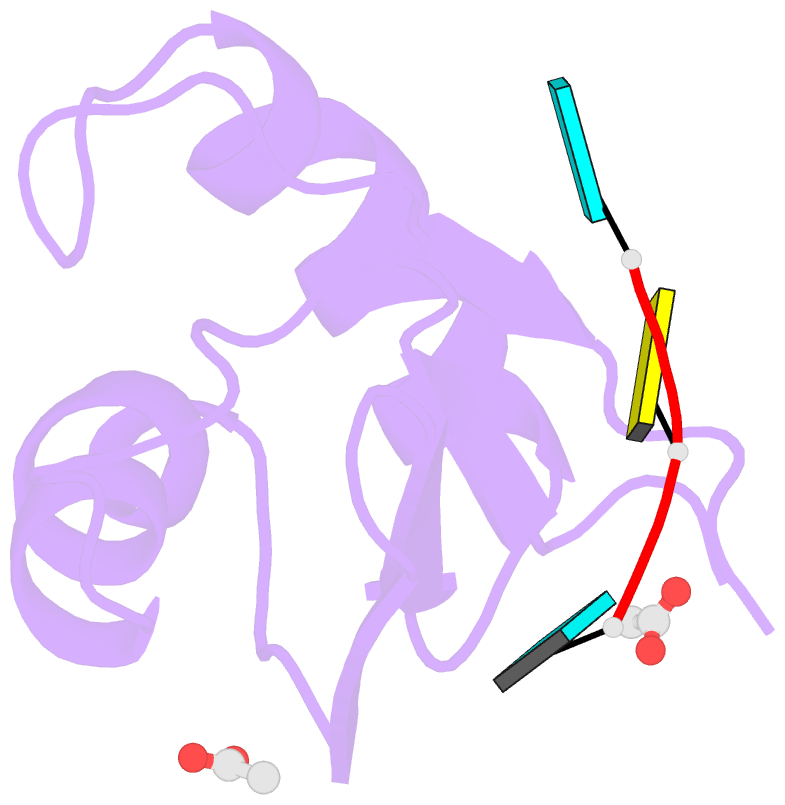
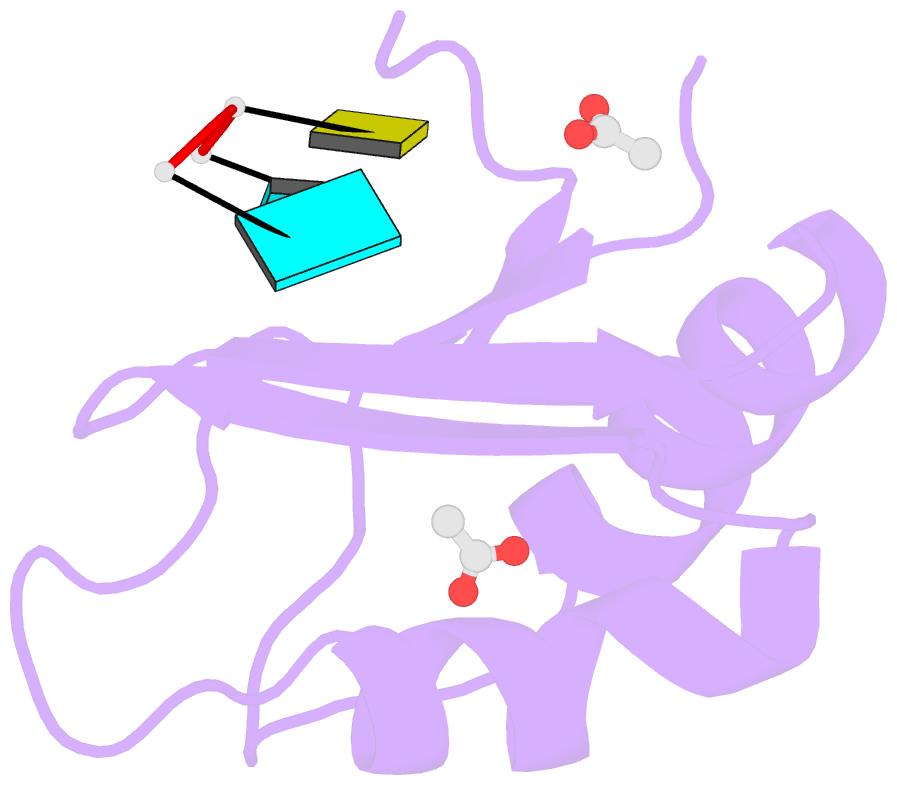
- Structural insights into cis element recognition of non- polyadenylated rnas by the nab3-rrm
- Reference
- Lunde BM, Horner M, Meinhart A (2011): "Structural Insights Into Cis Element Recognition of Non-Polyadenylated Rnas by the Nab3-Rrm." Nucleic Acids Res., 39, 337. doi: 10.1093/NAR/GKQ751.
- Abstract
- Transcription termination of non-polyadenylated RNAs in Saccharomyces cerevisiae occurs through the action of the Nrd1-Nab3-Sen1 complex. Part of the decision to terminate via this pathway occurs via direct recognition of sequences within the nascent transcript by RNA recognition motifs (RRMs) within Nrd1 and Nab3. Here we present the 1.6 Å structure of Nab3-RRM bound to its UCUU recognition sequence. The crystal structure reveals clear density for a UCU trinucleotide and a fourth putative U binding site. Nab3-RRM establishes a clear preference for the central cytidine of the UCUU motif, which forms pseudo-base pairing interactions primarily through hydrogen bonds to main chain atoms and one serine hydroxyl group. Specificity for the flanking uridines is less defined; however, binding experiments confirm that these residues are also important for high affinity binding. Comparison of the Nab3-RRM to other structures of RRMs bound to polypyrimidine RNAs showed that this mode of recognition is similar to what is observed for the polypyrimidine-tract binding RRMs, and that the serine residue involved in pseudo-base pairing is only found in RRMs that bind to polypyrimidine RNAs that contain a cytosine base, suggesting a possible mechanism for discriminating between cytosine and uracil bases in RRMs that bind to polypyrimidine-containing RNA.