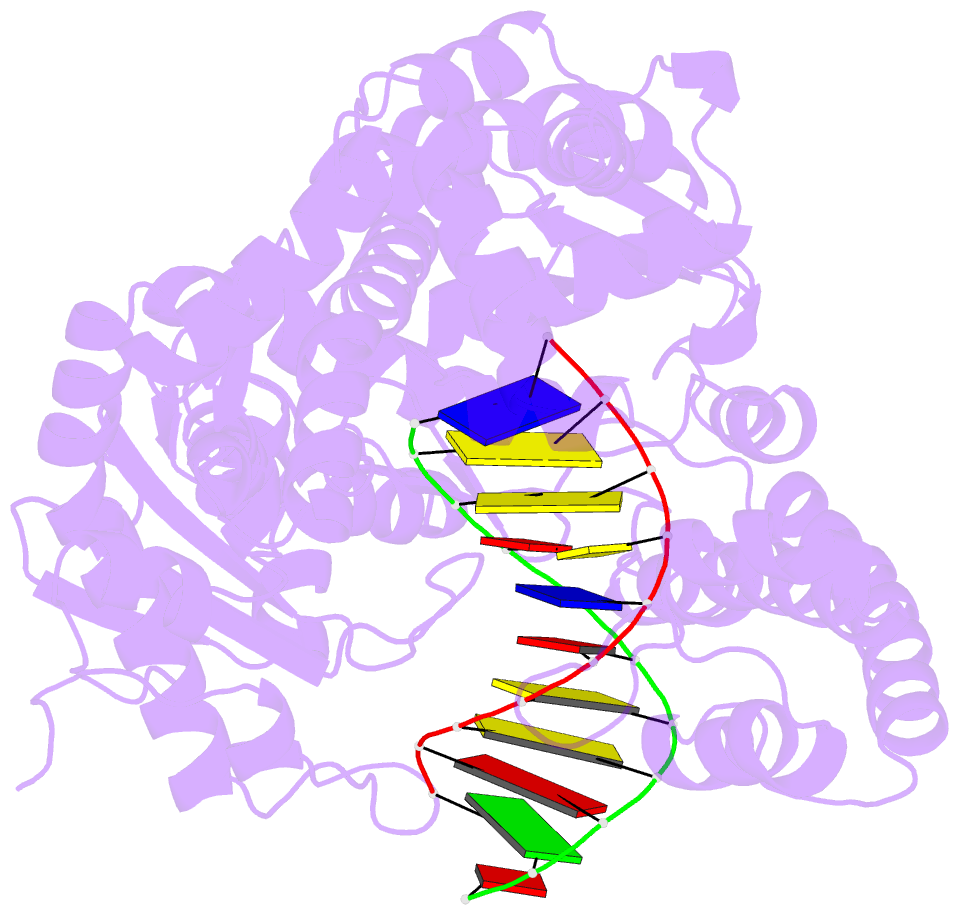
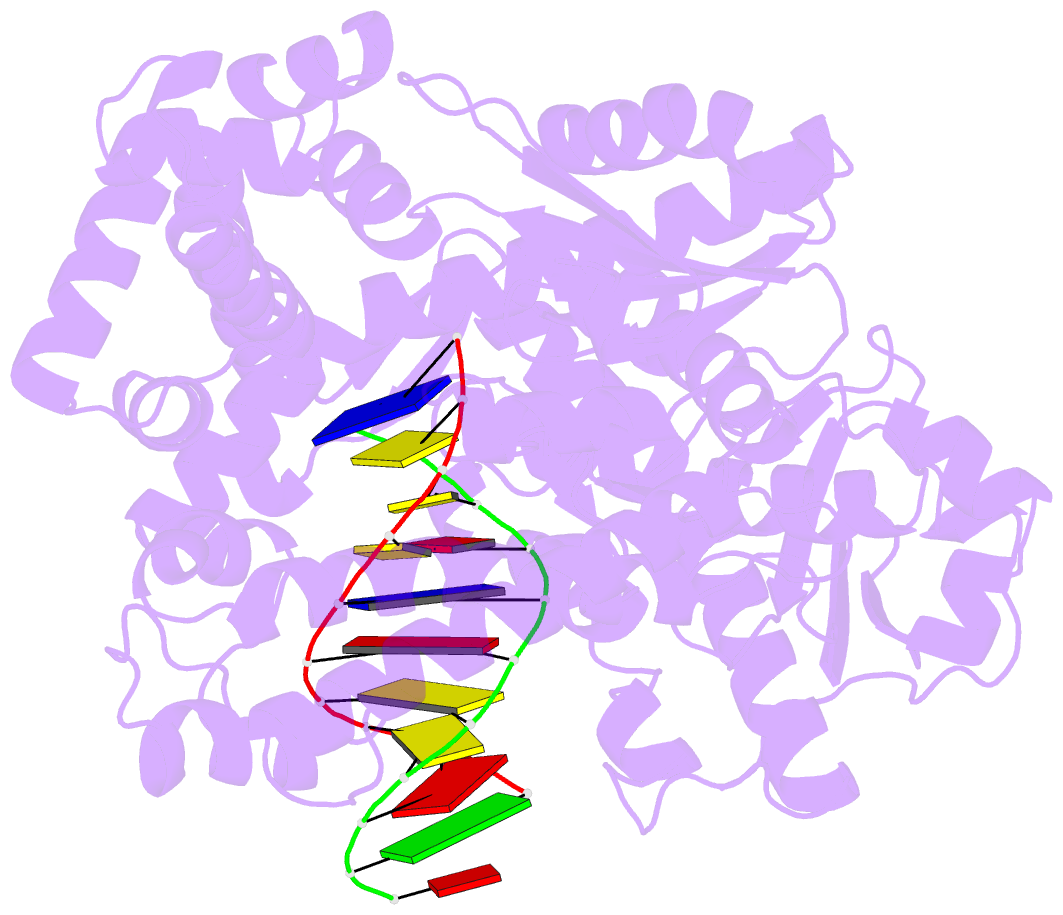
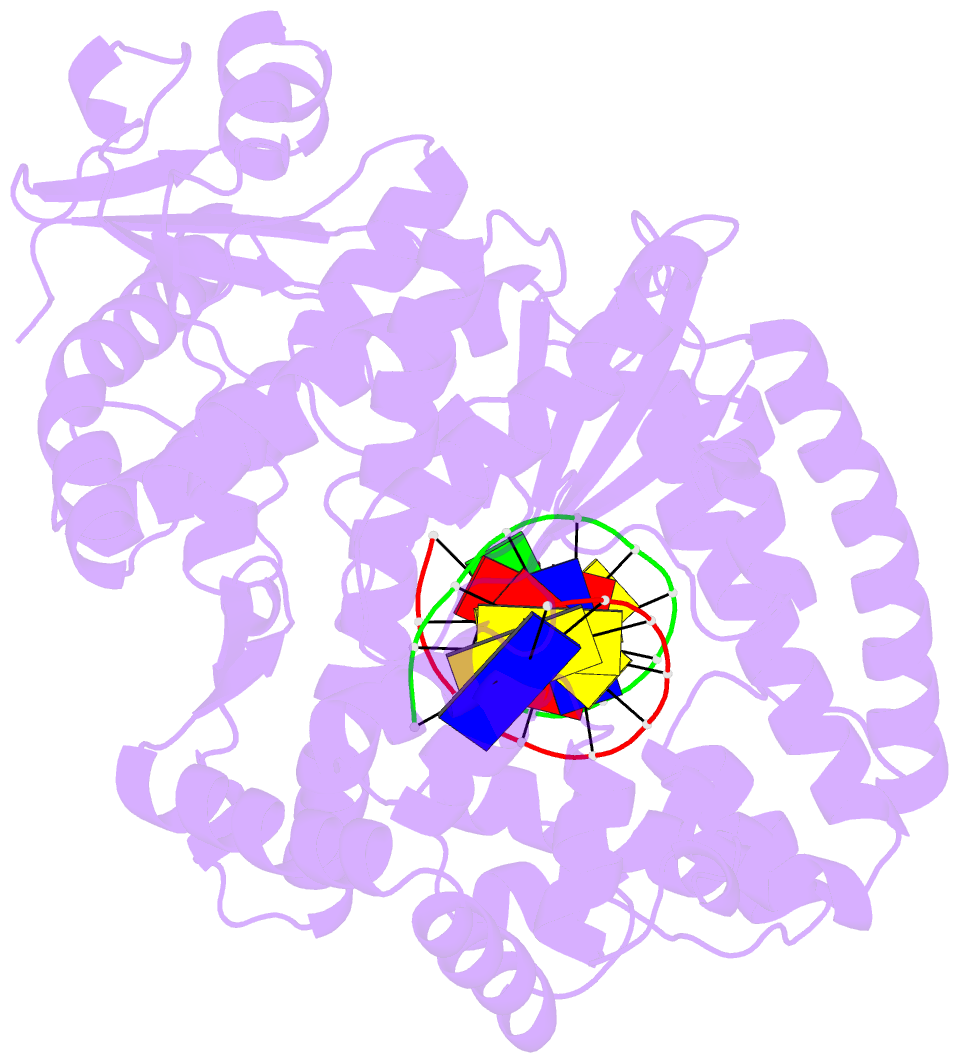
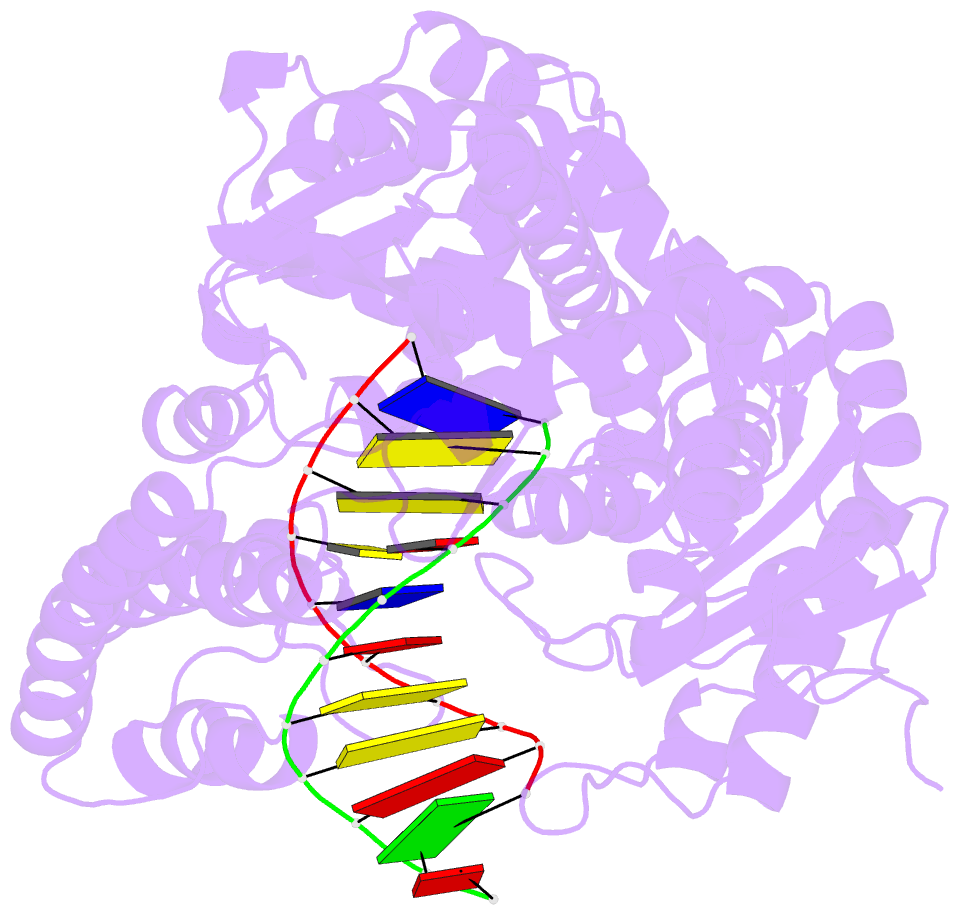
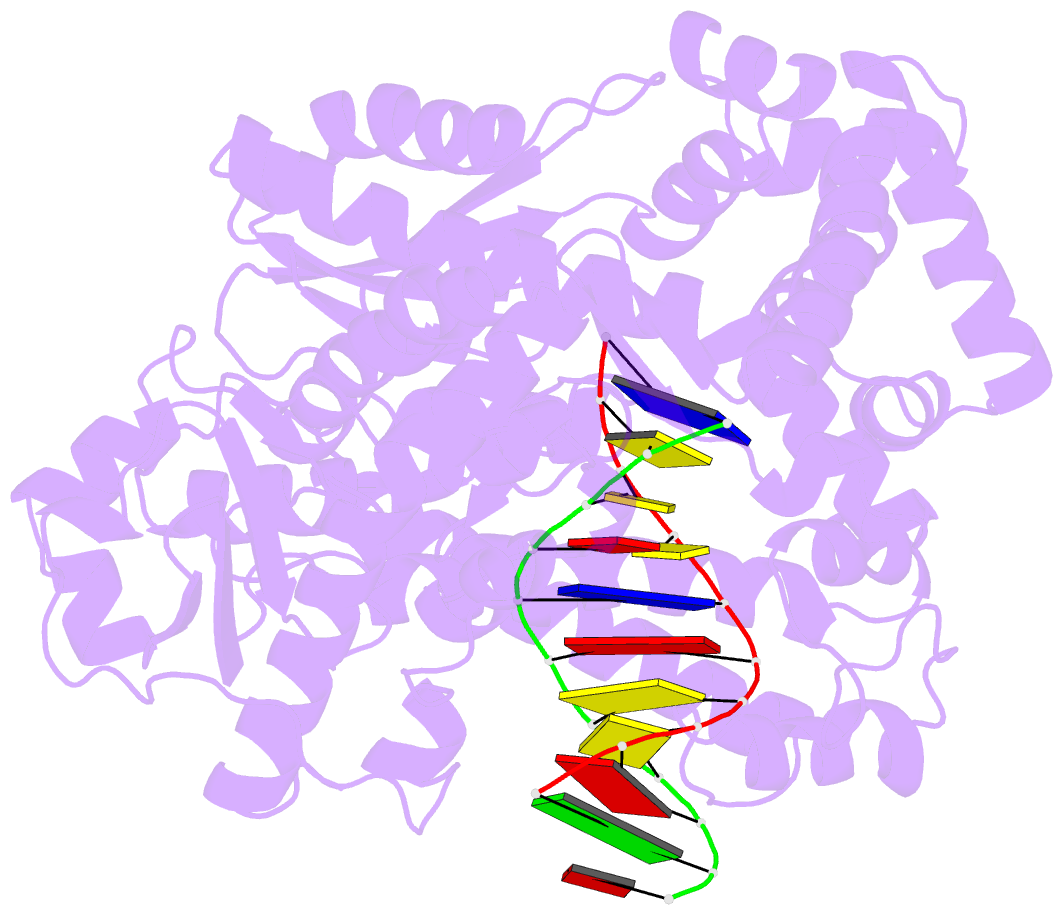
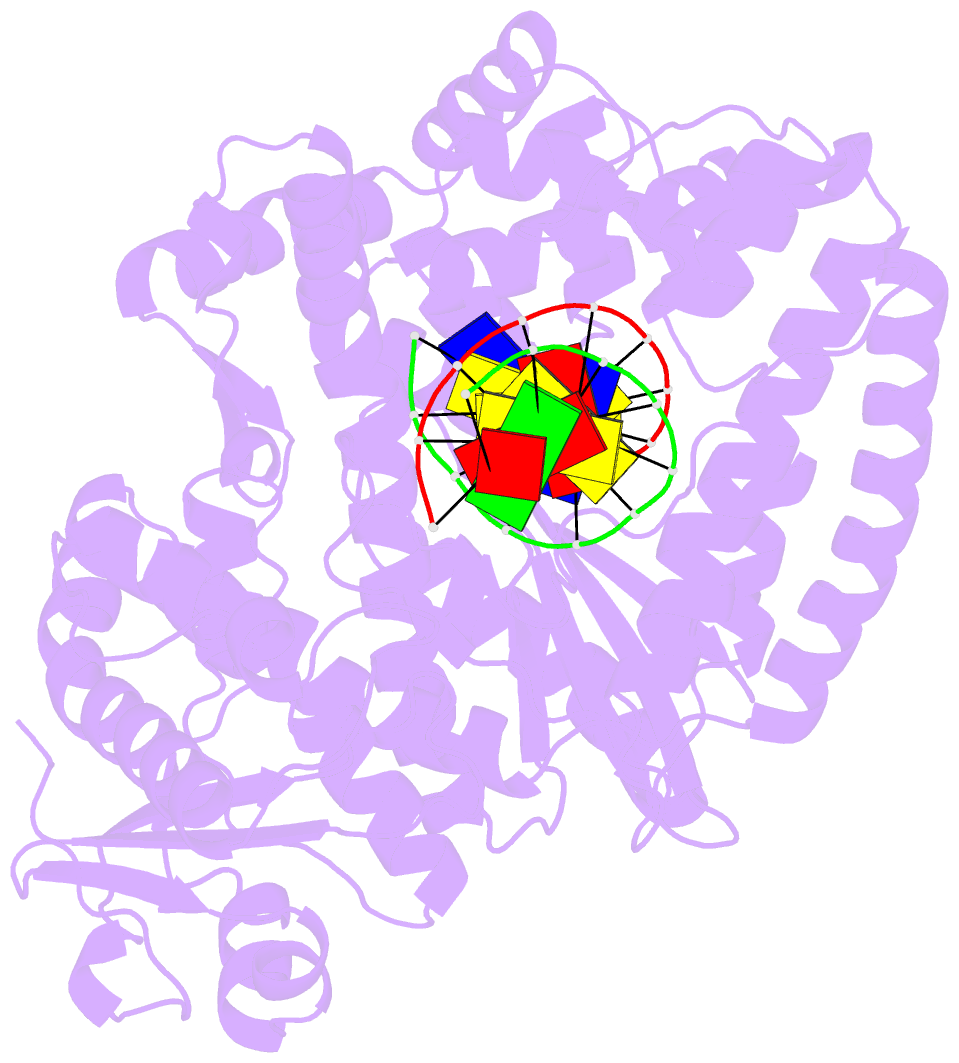
Summary information and primary citation
- PDB-id
- 2xo7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.85 Å)
- Summary
- Crystal structure of a da:o-allylhydroxylamine-dc basepair in complex with fragment DNA polymerase i from bacillus stearothermophilus
- Reference
- Muenzel M, Lercher L, Mueller M, Carell T (2010): "Chemical Discrimination between Dc and 5Medc Via Their Hydroxylamine Adducts." Nucleic Acids Res., 38, E192. doi: 10.1093/NAR/GKQ724.
- Abstract
- The presence of the methylated nucleobase (5Me)dC in CpG islands is a key factor that determines gene silencing. False methylation patterns are responsible for deteriorated cellular development and are a hallmark of many cancers. Today genes can be sequenced for the content of (5Me)dC only with the help of the bisulfite reagent, which is based exclusively on chemical reactivity differences established by the additional methyl group. Despite intensive optimization of the bisulfite protocol, the method still has specificity problems. Most importantly ∼95% of the DNA analyte is degraded during the analysis procedure. We discovered that the reagent O-allylhydroxylamine is able to discriminate between dC and (5Me)dC. The reagent, in contrast to bisulfite, does not exploit reactivity differences but gives directly different reaction products. The reagent forms a stable mutagenic adduct with dC, which can exist in two states (E versus Z). In case of dC the allylhydroxylamine adduct switches into the E-isomeric form, which generates dC to dT transition mutations that can easily be detected by established methods. Significantly, the (5Me)dC-adduct adopts exclusively the Z-isomeric form, which causes the polymerase to stop. O-allylhydroxylamine does allow differentiation between dC and (5Me)dC with high accuracy, leading towards a novel and mild chemistry for methylation analysis.