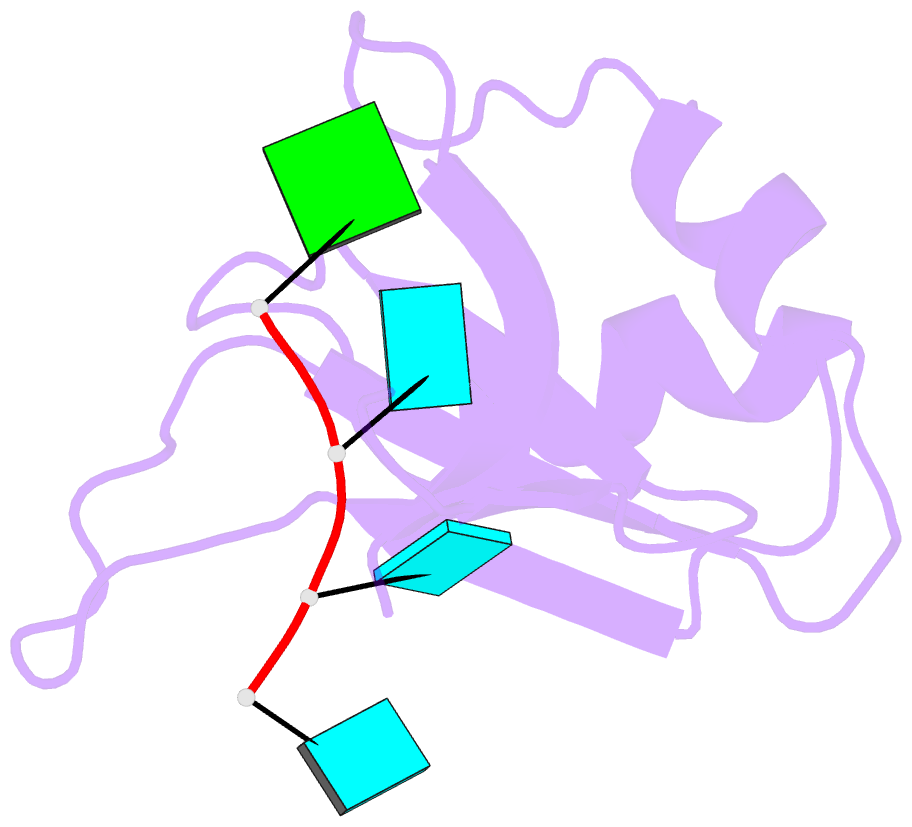
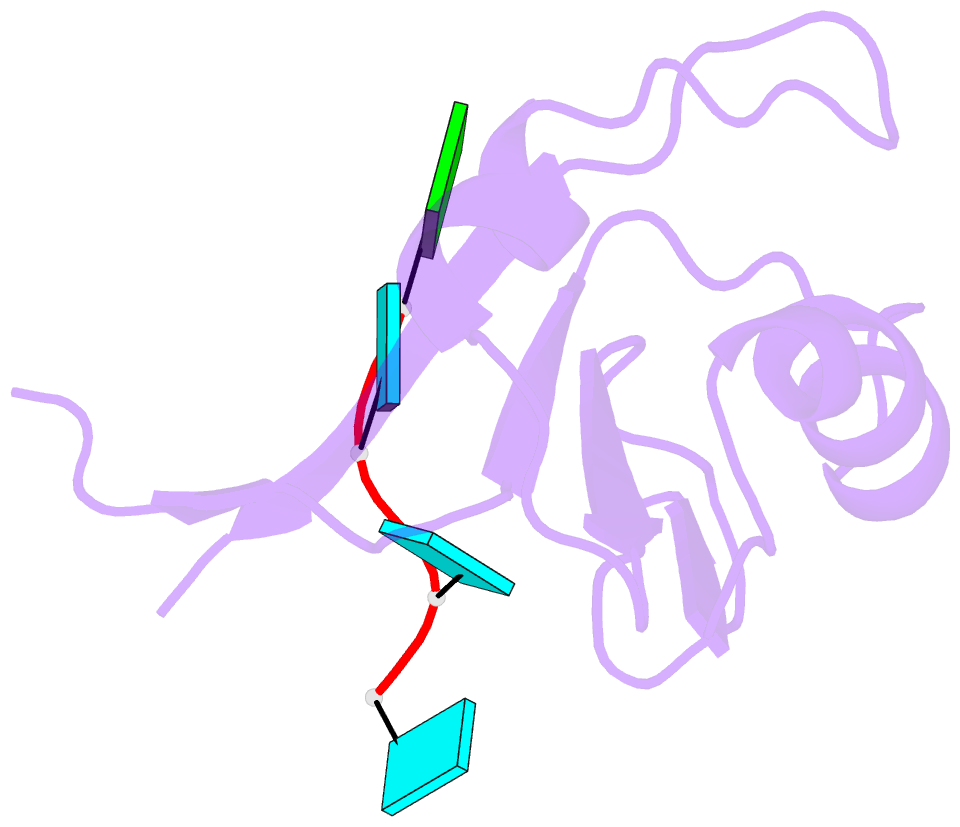
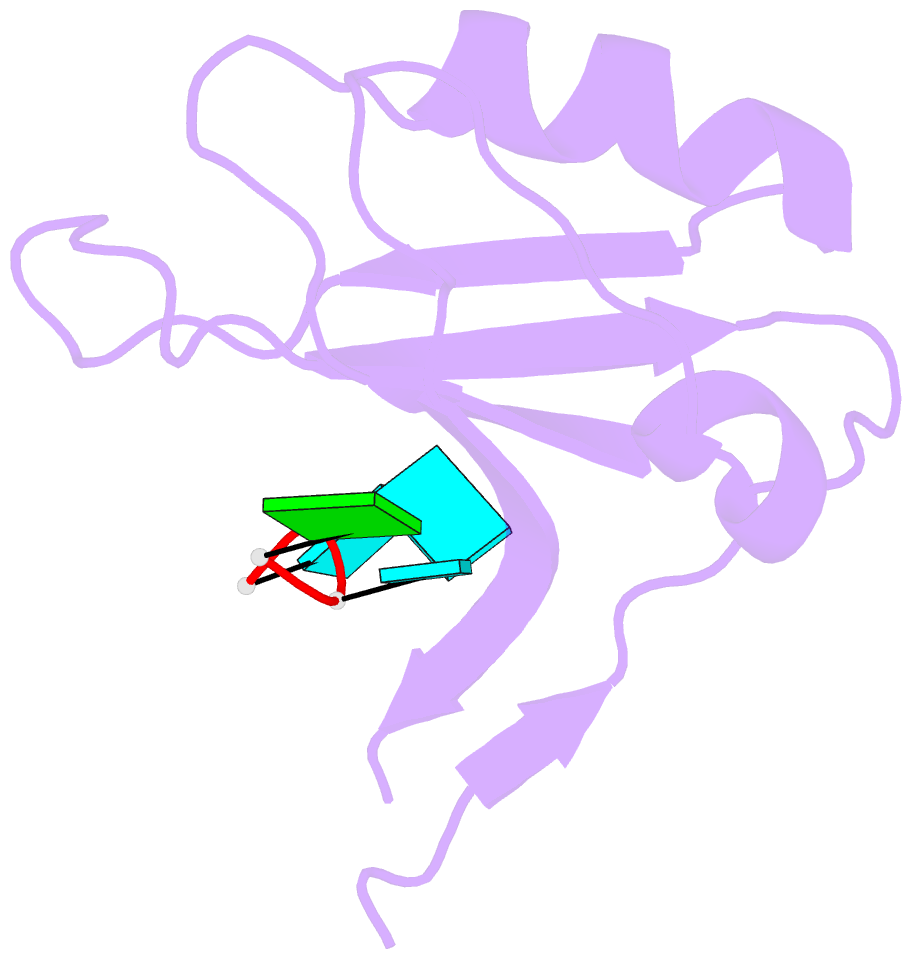
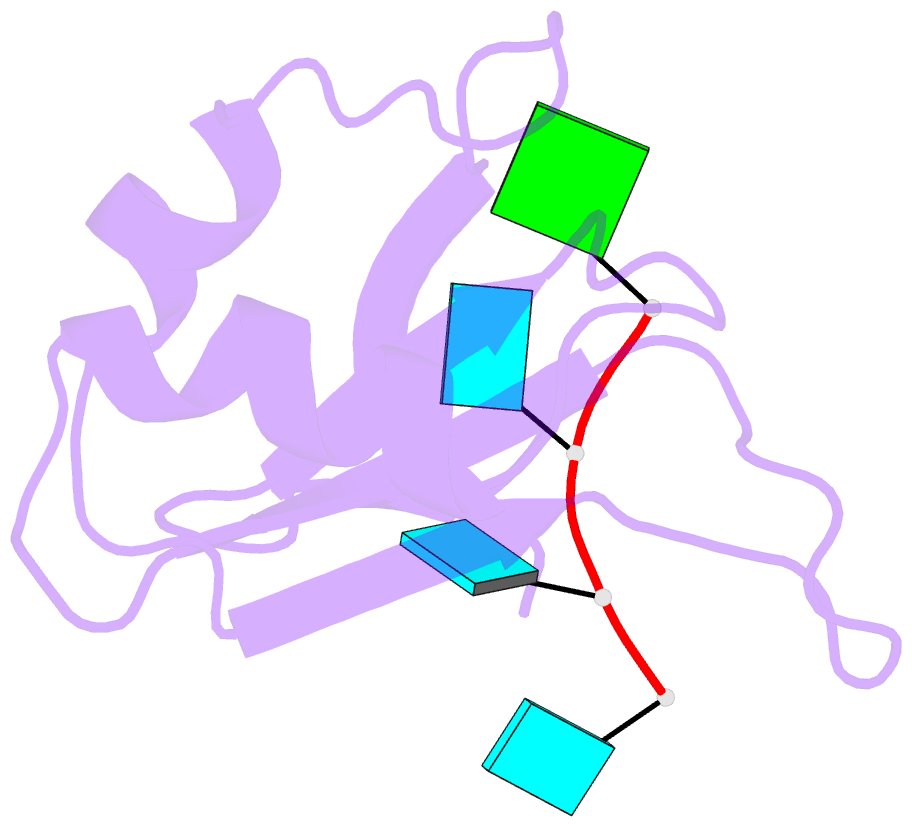
Summary information and primary citation
- PDB-id
- 2xs7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (1.45 Å)
- Summary
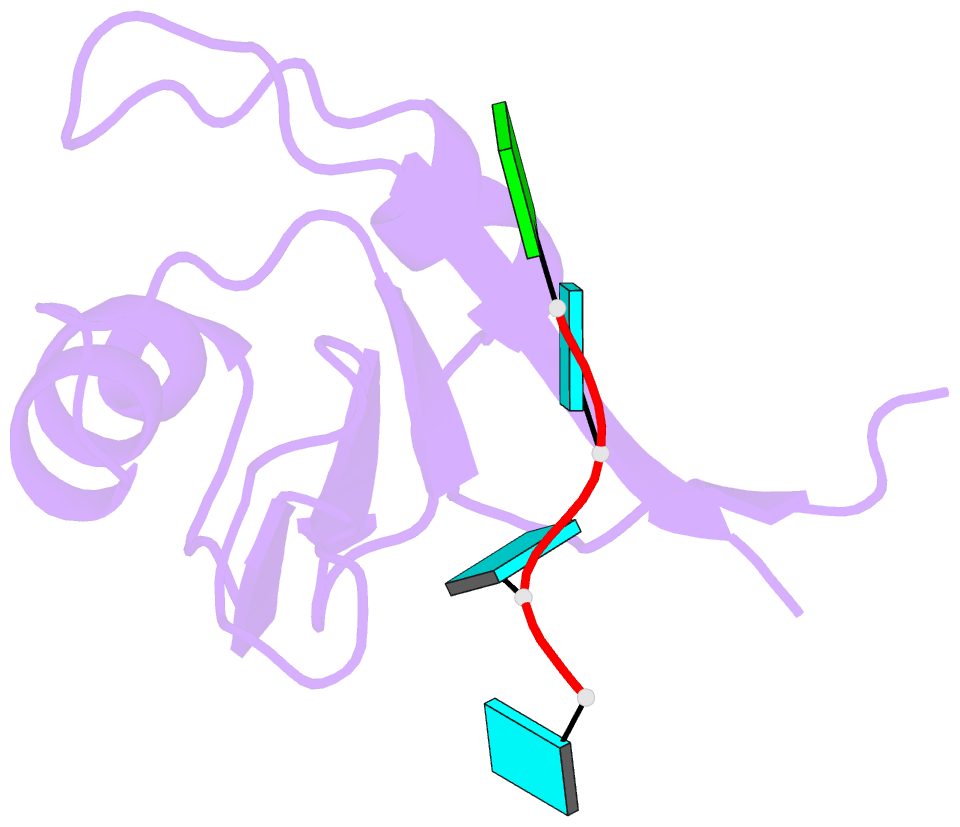
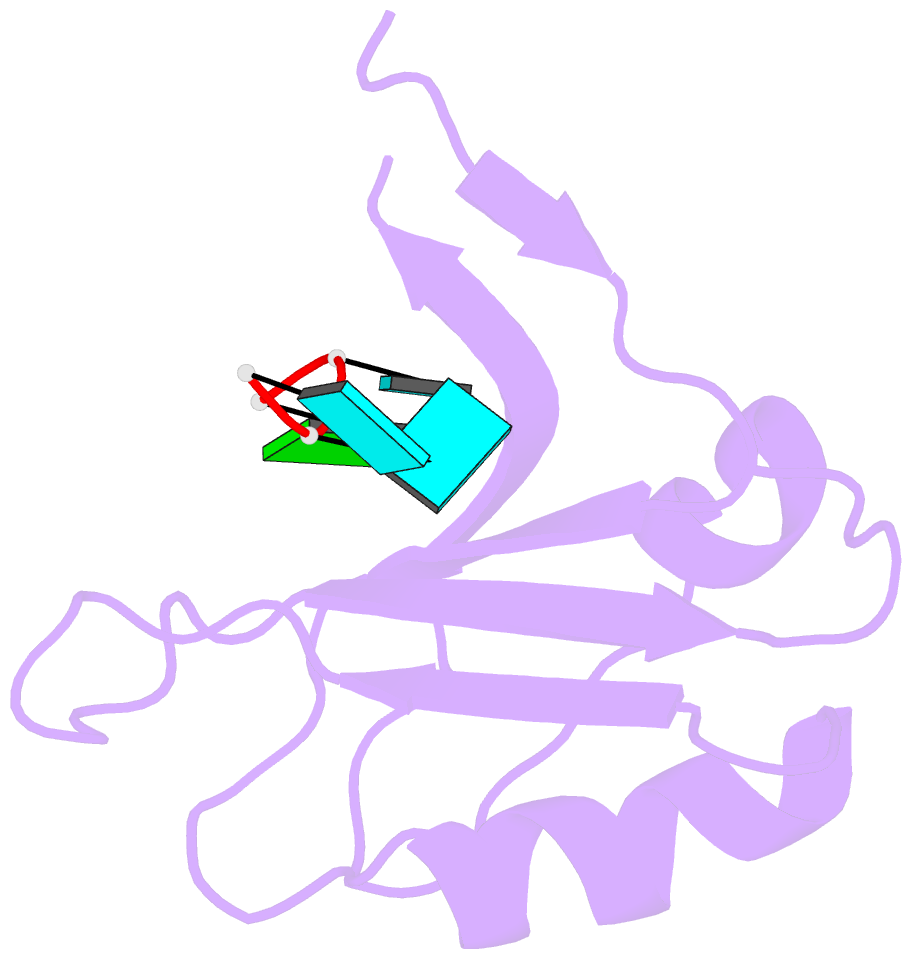
- Crystal structure of the rrm domain of mouse deleted in azoospermia- like in complex with sycp3 RNA, uuguuu
- Reference
- Jenkins HT, Edwards TA (2011): "Kinked Beta-Strands Mediate High-Affinity Recognition of Mrna Targets by the Germ-Cell Regulator Dazl." Proc.Natl.Acad.Sci.USA, 108, 18266. doi: 10.1073/PNAS.1105211108.
- Abstract
- A defect in germ-cell (sperm and oocyte) development is the leading cause of male and female infertility. Control of translation through the binding of deleted in azoospermia (DAZ)-like (DAZL) to the 3'-UTRs of mRNAs, via a highly conserved RNA recognition motif (RRM), has been shown to be essential in germ-cell development. Crystal structures of the RRM from murine DAZL (Dazl), both alone and in complex with RNA sequences from the 3'-UTRs of mRNAs regulated by Dazl, reveal high-affinity sequence-specific recognition of a GUU triplet involving an extended, kinked, pair of β-strands. Recognition of the GUU triplet is maintained, whereas the identity and position of bases flanking this triplet varies. The Dazl RRM is thus able to recognize GUU triplets in different sequence contexts. Mutation of bases within the GUU triplet reduces the affinity of binding. Together with the demonstration that multiple Dazl RRMs can bind to a single RNA containing multiple GUU triplets, these structures suggest that the number of DAZL molecules bound to GUU triplets in the 3'-UTR provides a method for modulating the translation of a target RNA. The conservation of RNA binding and structurally important residues between members of the DAZ family, together with the demonstration that mutation of these residues severely impairs RNA binding, indicate that the mode of RNA binding revealed by these structures is conserved in proteins essential for gamete development from flies to humans.