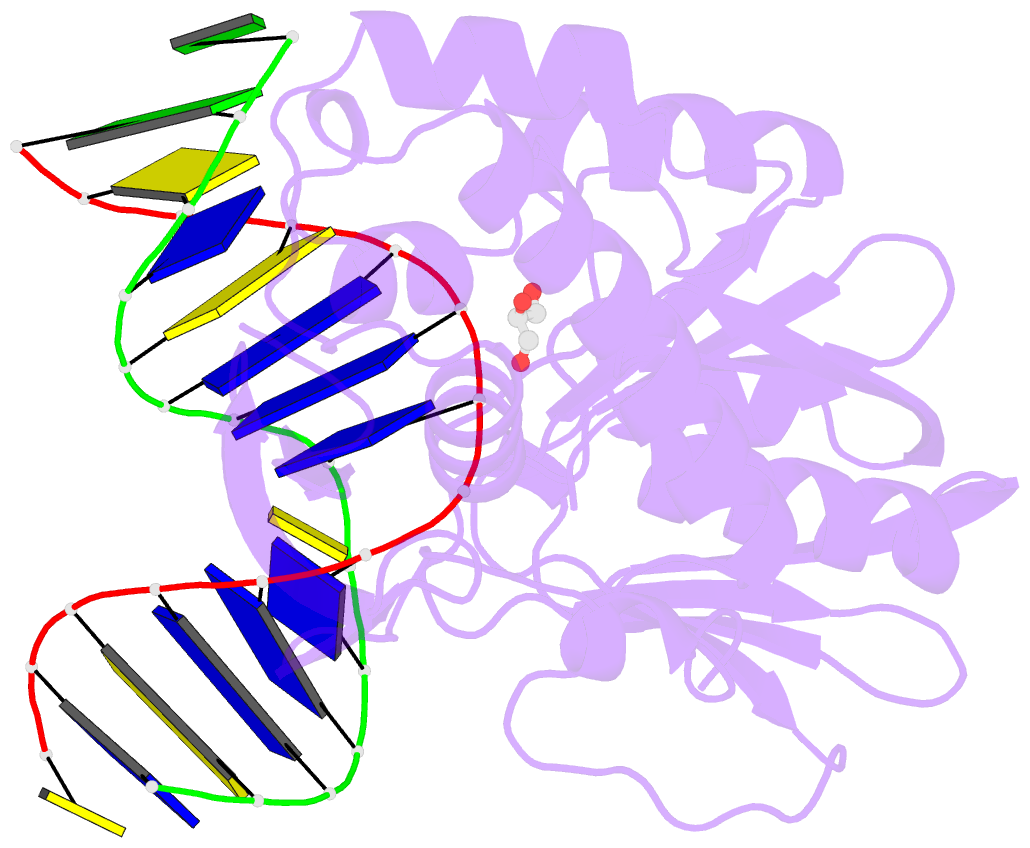
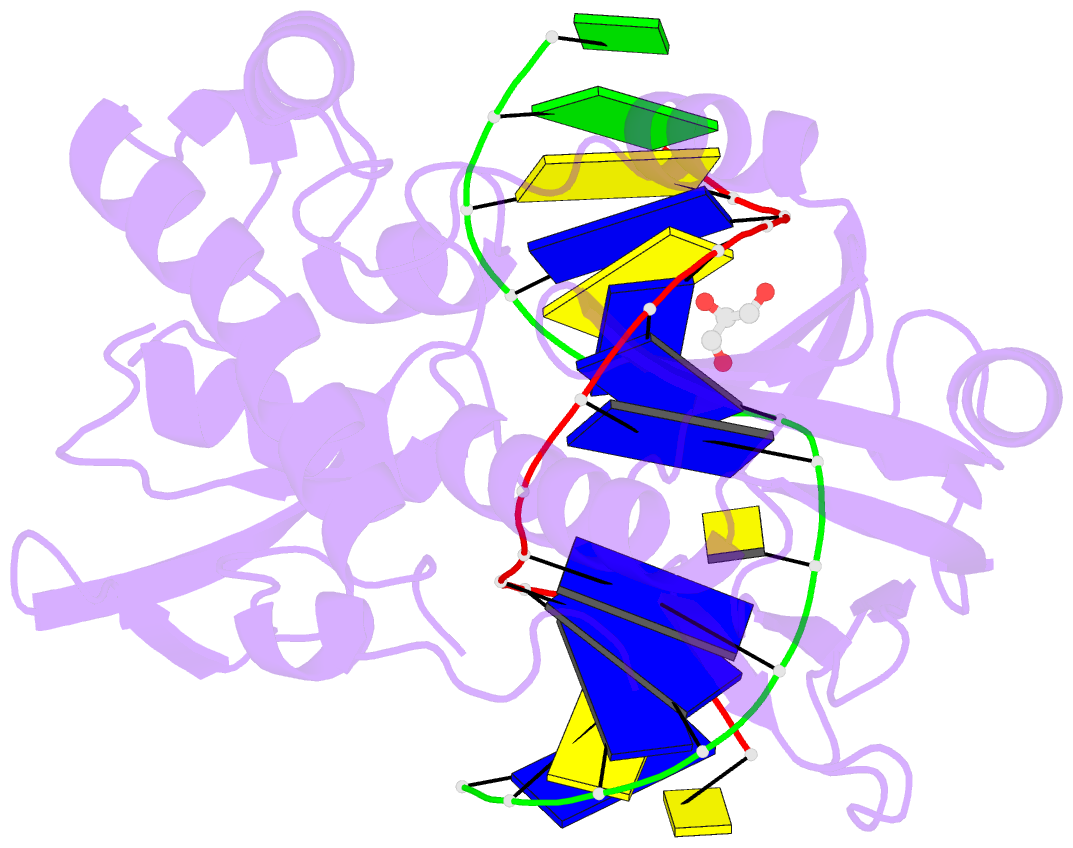
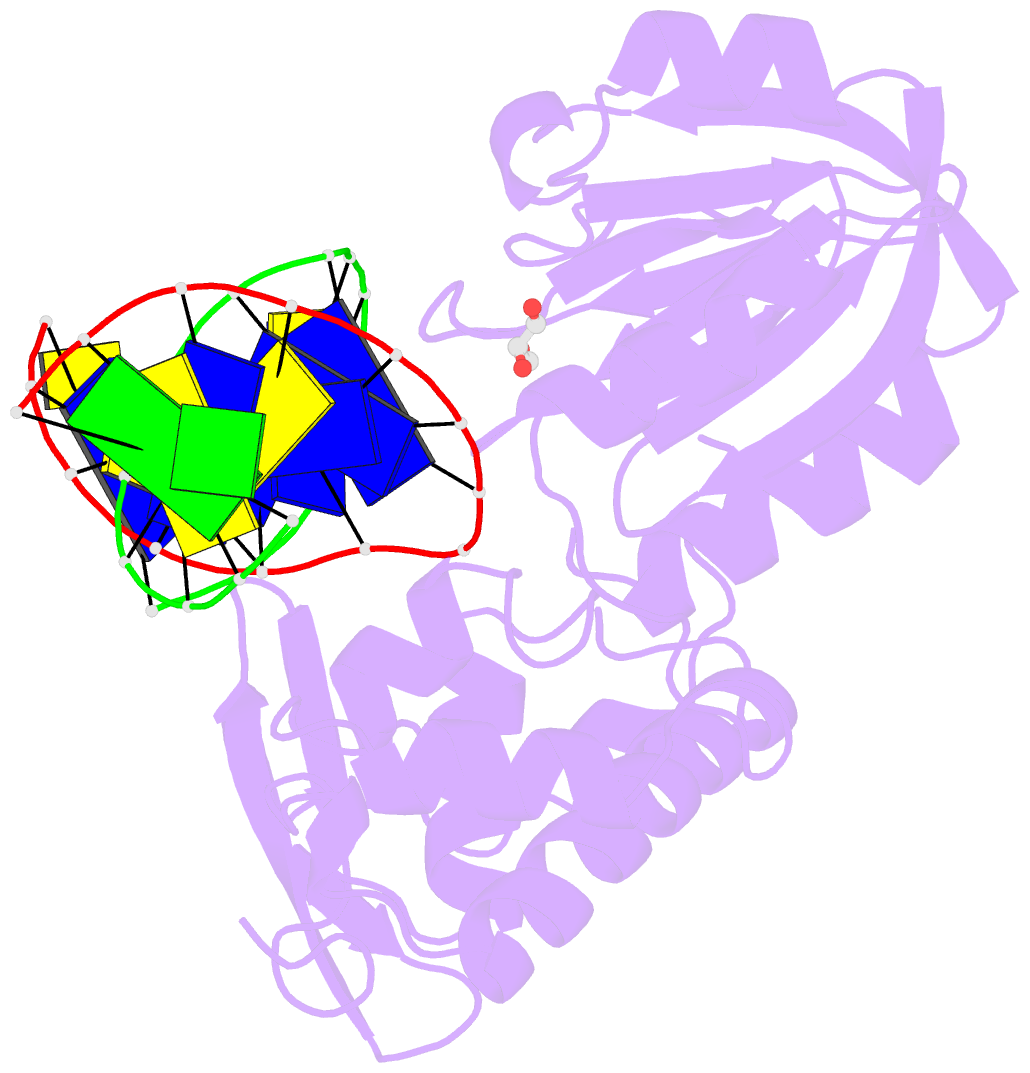
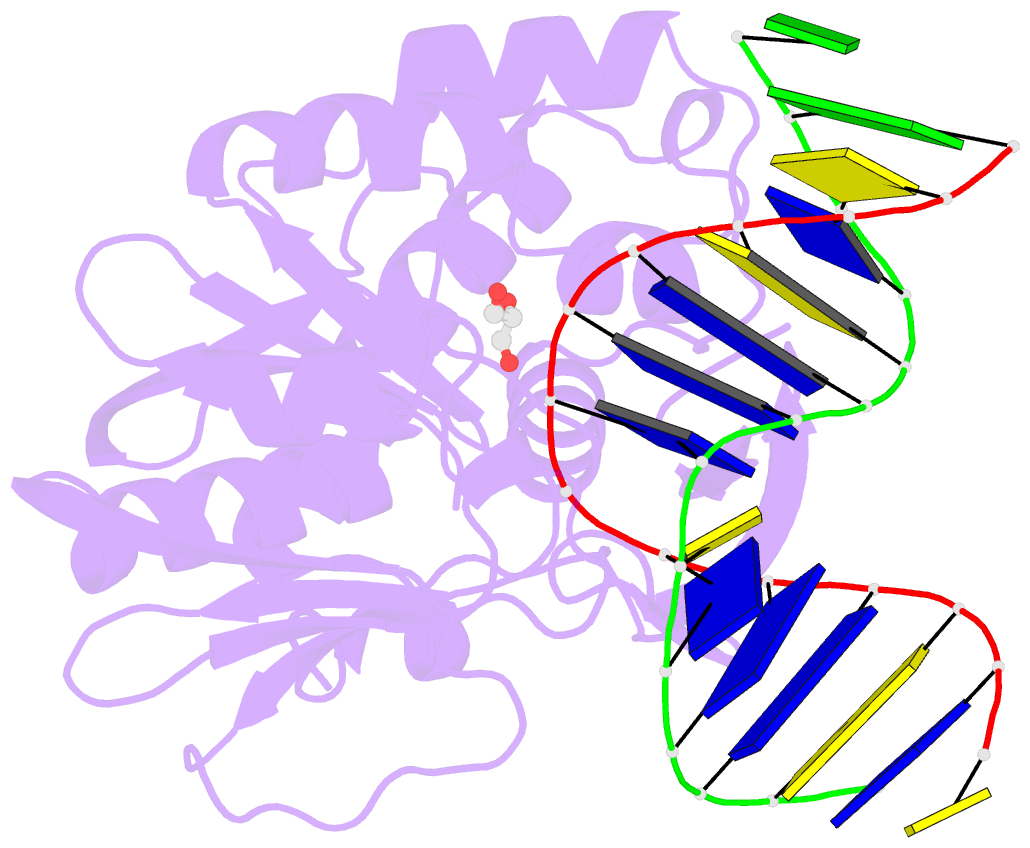
Summary information and primary citation
- PDB-id
- 2xzu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.82 Å)
- Summary
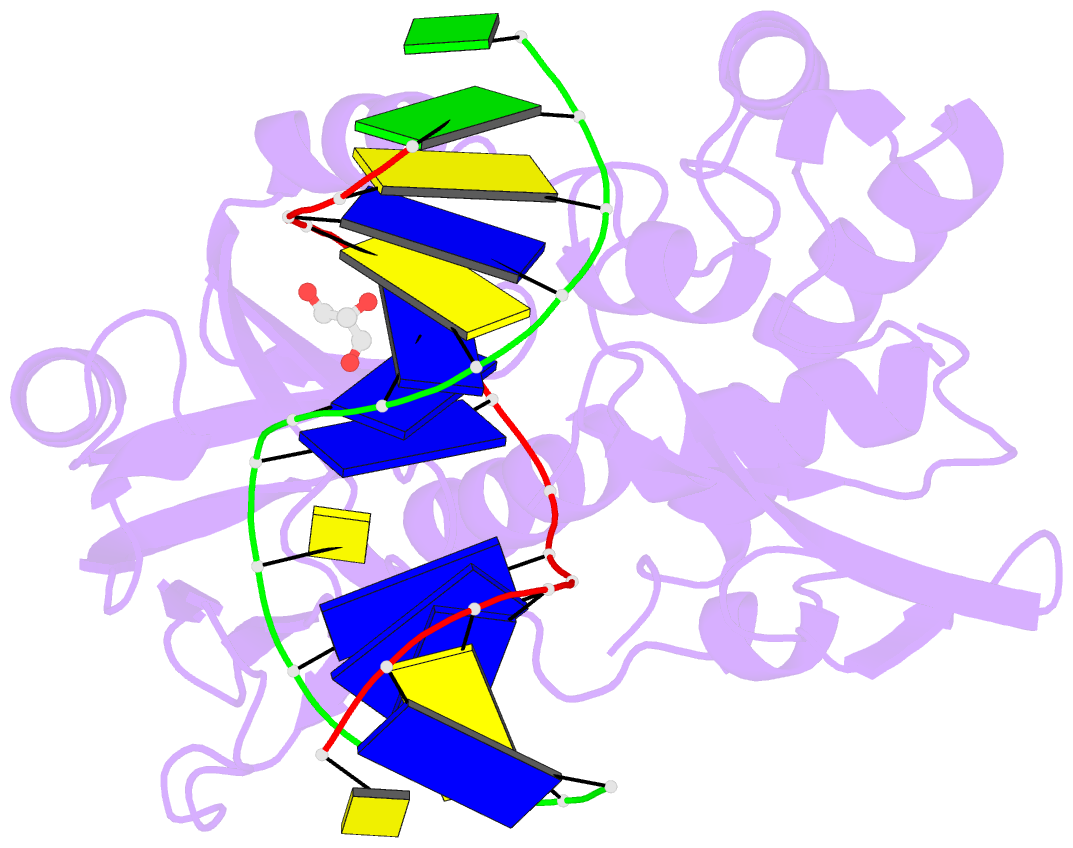
- Crystal structure of a complex between the wild-type lactococcus lactis fpg (mutm) and an oxidized pyrimidine containing DNA at 310k
- Reference
- Le Bihan YV, Izquierdo MA, Coste F, Aller P, Culard F, Gehrke TH, Essalhi K, Carrel T, Castaing B (2011): "5-Hydroxy-5-Methylhydantoin DNA Lesion, a Molecular Trap for DNA Glycosylases." Nucleic Acids Res., 39, 6277. doi: 10.1093/NAR/GKR215.
- Abstract
- DNA base-damage recognition in the base excision repair (BER) is a process operating on a wide variety of alkylated, oxidized and degraded bases. DNA glycosylases are the key enzymes which initiate the BER pathway by recognizing and excising the base damages guiding the damaged DNA through repair synthesis. We report here biochemical and structural evidence for the irreversible entrapment of DNA glycosylases by 5-hydroxy-5-methylhydantoin, an oxidized thymine lesion. The first crystal structure of a suicide complex between DNA glycosylase and unrepaired DNA has been solved. In this structure, the formamidopyrimidine-(Fapy) DNA glycosylase from Lactococcus lactis (LlFpg/LlMutM) is covalently bound to the hydantoin carbanucleoside-containing DNA. Coupling a structural approach by solving also the crystal structure of the non-covalent complex with site directed mutagenesis, this atypical suicide reaction mechanism was elucidated. It results from the nucleophilic attack of the catalytic N-terminal proline of LlFpg on the C5-carbon of the base moiety of the hydantoin lesion. The biological significance of this finding is discussed.