Summary information and primary citation
- PDB-id
- 2y35; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (3.2 Å)
- Summary
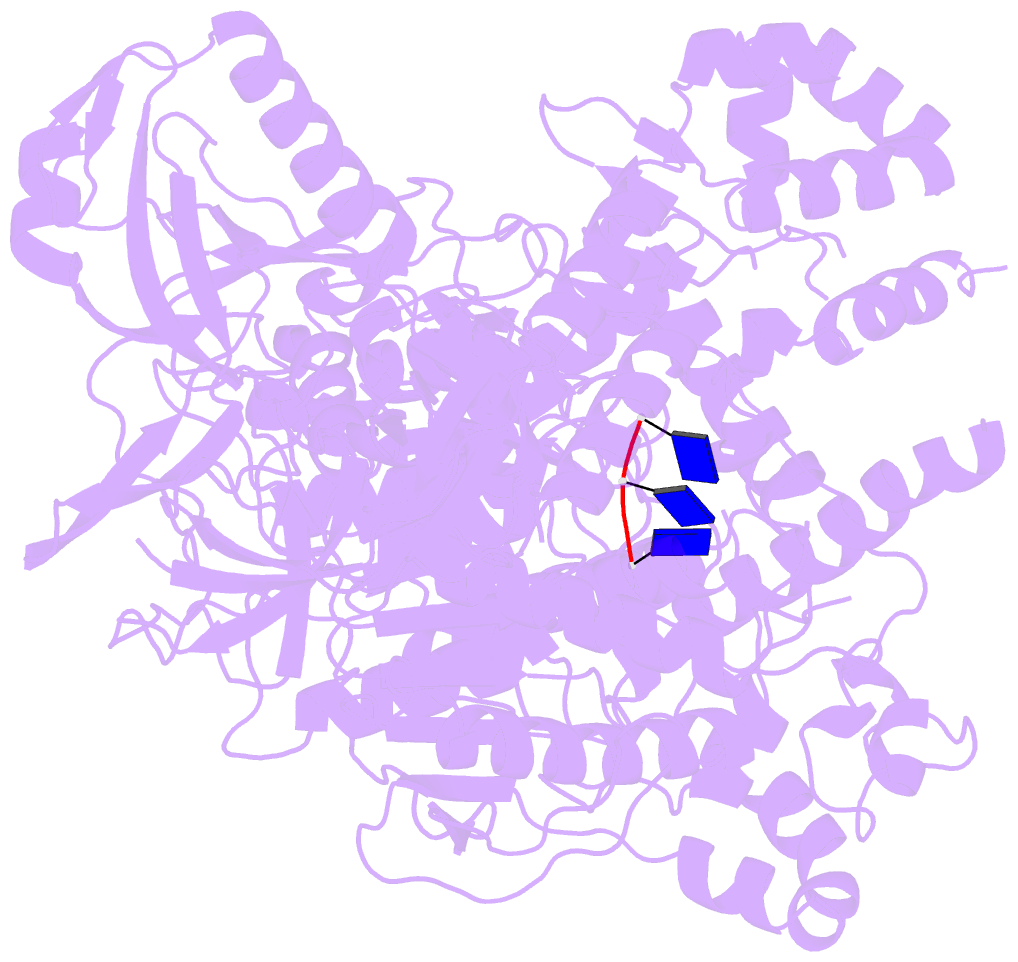
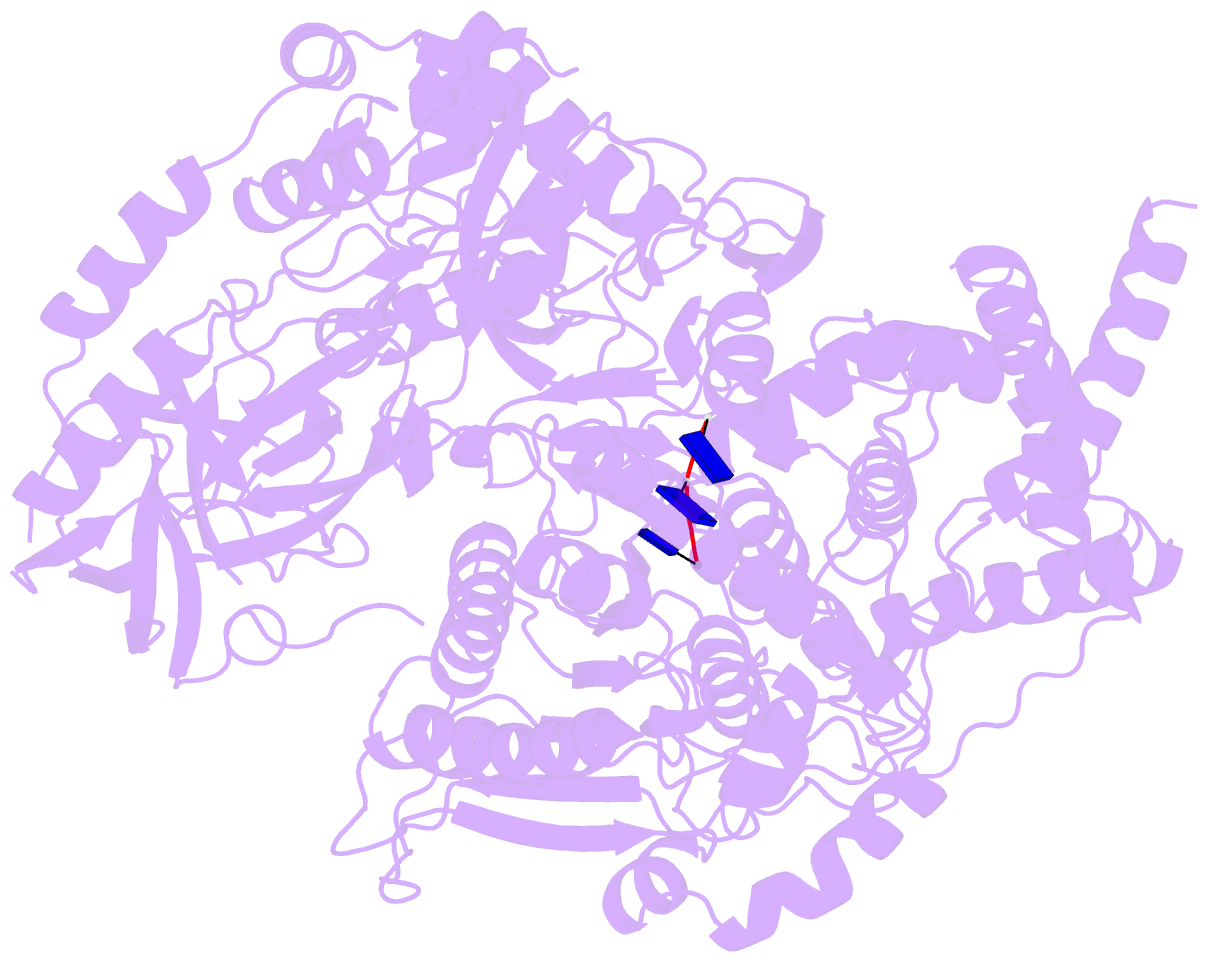
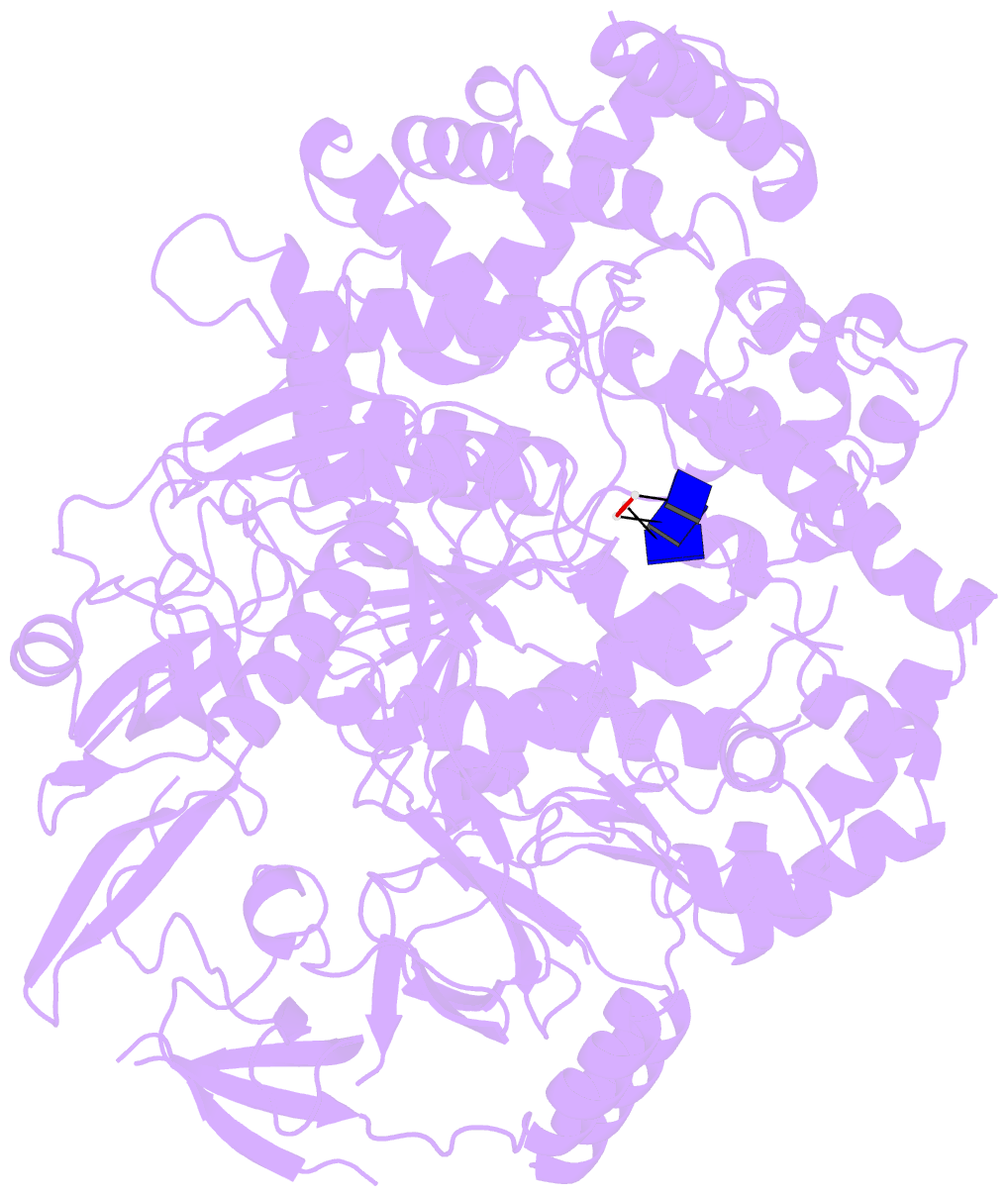
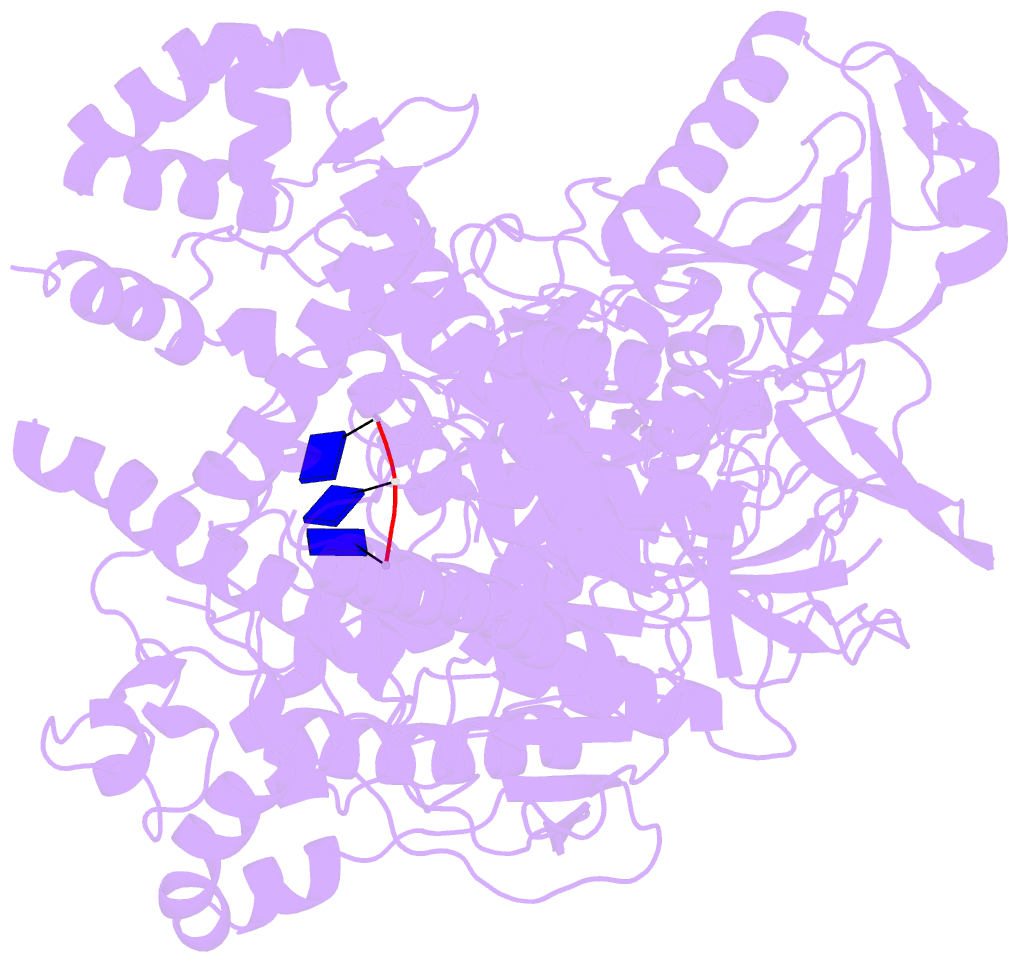
- Crystal structure of xrn1-substrate complex
- Reference
- Jinek M, Coyle SM, Doudna JA (2011): "Coupled 5' Nucleotide Recognition and Processivity in Xrn1-Mediated Mrna Decay." Mol.Cell, 41, 600. doi: 10.1016/J.MOLCEL.2011.02.004.
- Abstract
- Messenger RNA decay plays a central role in the regulation and surveillance of eukaryotic gene expression. The conserved multidomain exoribonuclease Xrn1 targets cytoplasmic RNA substrates marked by a 5' monophosphate for processive 5'-to-3' degradation by an unknown mechanism. Here, we report the crystal structure of an Xrn1-substrate complex. The single-stranded substrate is held in place by stacking of the 5'-terminal trinucleotide between aromatic side chains while a highly basic pocket specifically recognizes the 5' phosphate. Mutations of residues involved in binding the 5'-terminal nucleotide impair Xrn1 processivity. The substrate recognition mechanism allows Xrn1 to couple processive hydrolysis to duplex melting in RNA substrates with sufficiently long single-stranded 5' overhangs. The Xrn1-substrate complex structure thus rationalizes the exclusive specificity of Xrn1 for 5'-monophosphorylated substrates, ensuring fidelity of mRNA turnover, and posits a model for translocation-coupled unwinding of structured RNA substrates.