Summary information and primary citation
- PDB-id
- 2yh1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- NMR
- Summary
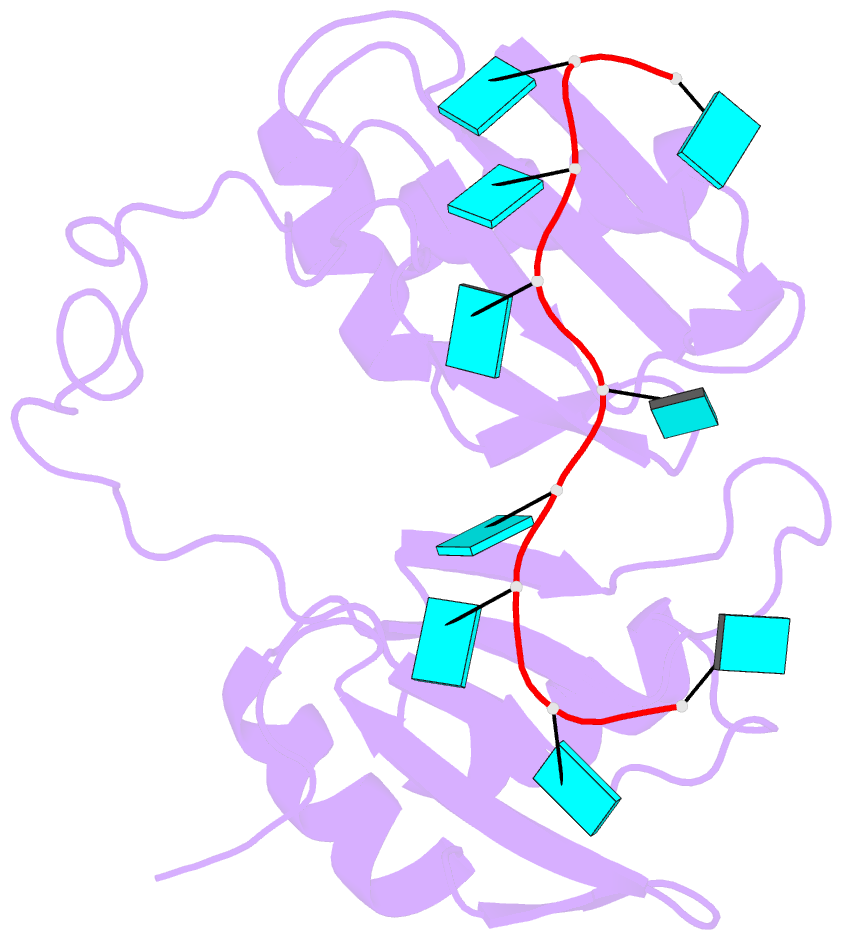
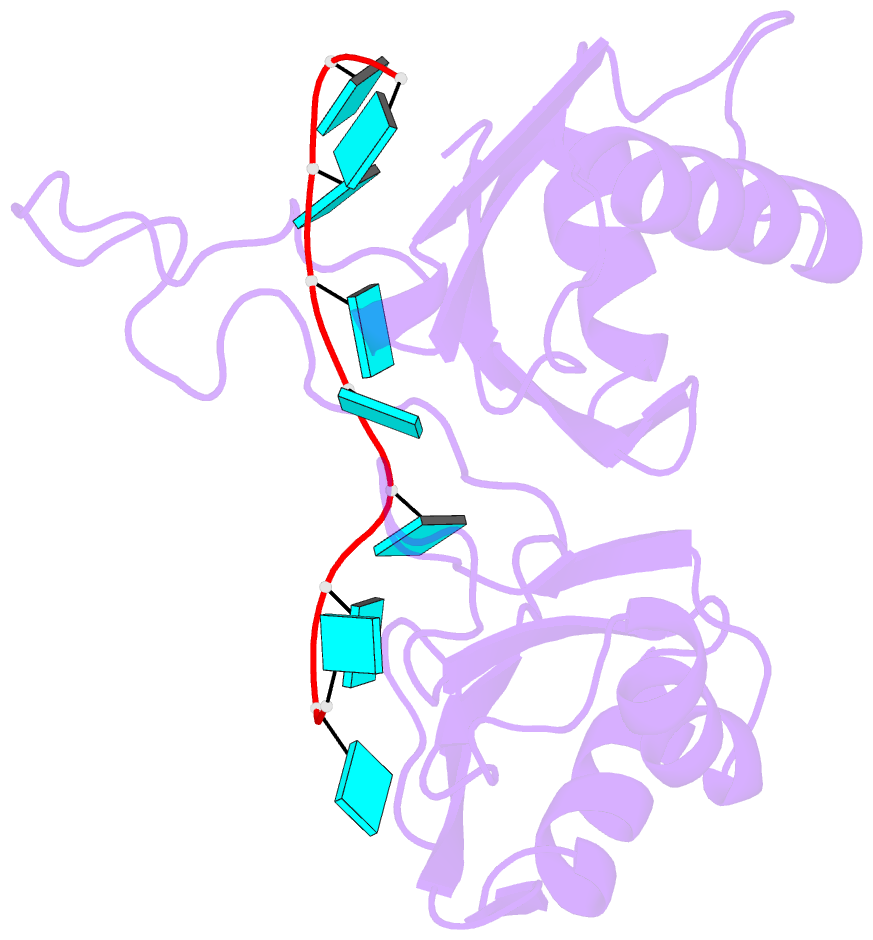
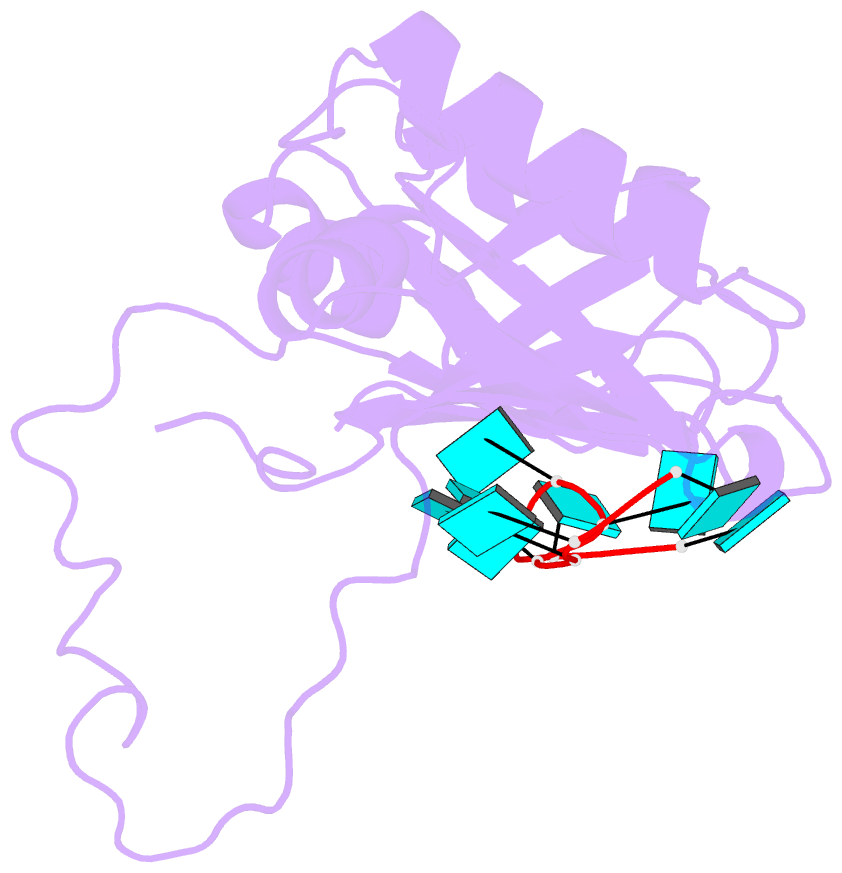
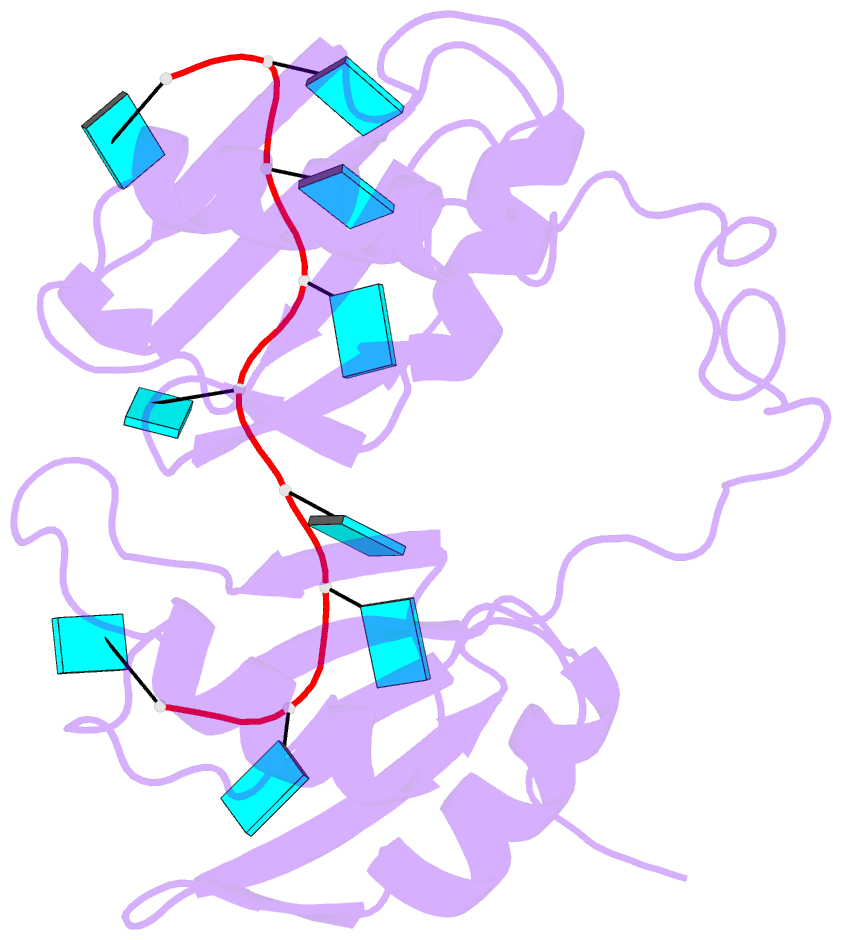
- Model of human u2af65 tandem rrm1 and rrm2 domains with eight-site uridine binding
- Reference
- Mackereth CD, Madl T, Bonnal S, Simon B, Zanier K, Gasch A, Rybin V, Valcarcel J, Sattler M (2011): "Multi-Domain Conformational Selection Underlies Pre-Mrna Splicing Regulation by U2Af." Nature, 475, 408. doi: 10.1038/NATURE10171.
- Abstract
- Many cellular functions involve multi-domain proteins, which are composed of structurally independent modules connected by flexible linkers. Although it is often well understood how a given domain recognizes a cognate oligonucleotide or peptide motif, the dynamic interaction of multiple domains in the recognition of these ligands remains to be characterized. Here we have studied the molecular mechanisms of the recognition of the 3'-splice-site-associated polypyrimidine tract RNA by the large subunit of the human U2 snRNP auxiliary factor (U2AF65) as a key early step in pre-mRNA splicing. We show that the tandem RNA recognition motif domains of U2AF65 adopt two remarkably distinct domain arrangements in the absence or presence of a strong (that is, high affinity) polypyrimidine tract. Recognition of sequence variations in the polypyrimidine tract RNA involves a population shift between these closed and open conformations. The equilibrium between the two conformations functions as a molecular rheostat that quantitatively correlates the natural variations in polypyrimidine tract nucleotide composition, length and functional strength to the efficiency to recruit U2 snRNP to the intron during spliceosome assembly. Mutations that shift the conformational equilibrium without directly affecting RNA binding modulate splicing activity accordingly. Similar mechanisms of cooperative multi-domain conformational selection may operate more generally in the recognition of degenerate nucleotide or amino acid motifs by multi-domain proteins.