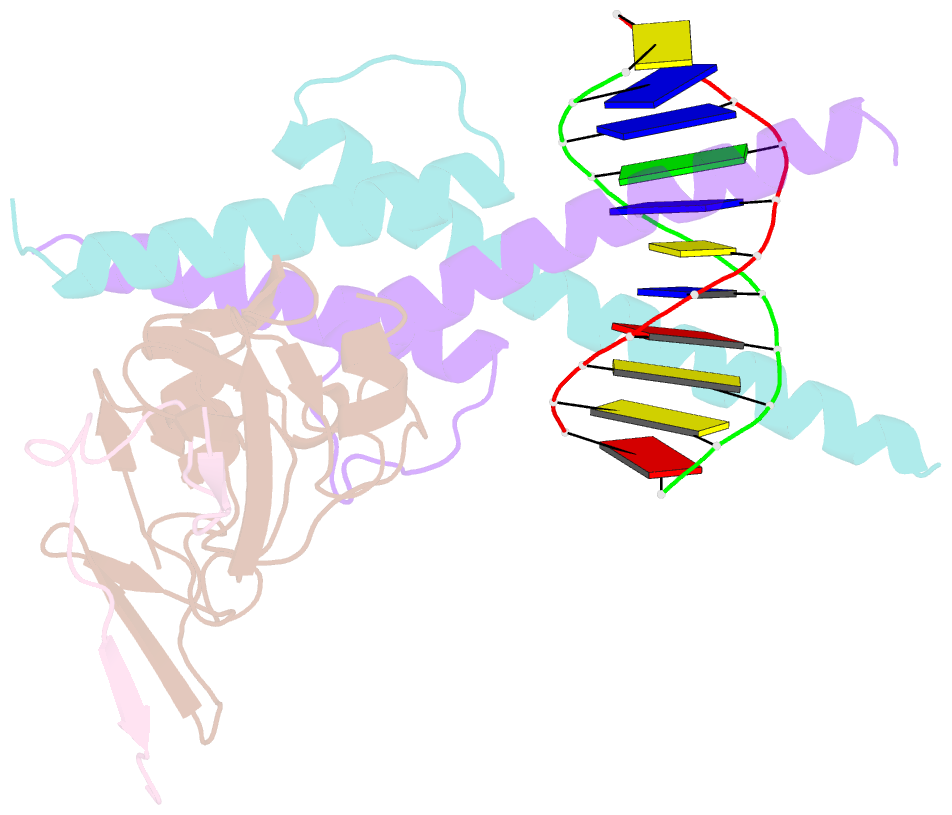
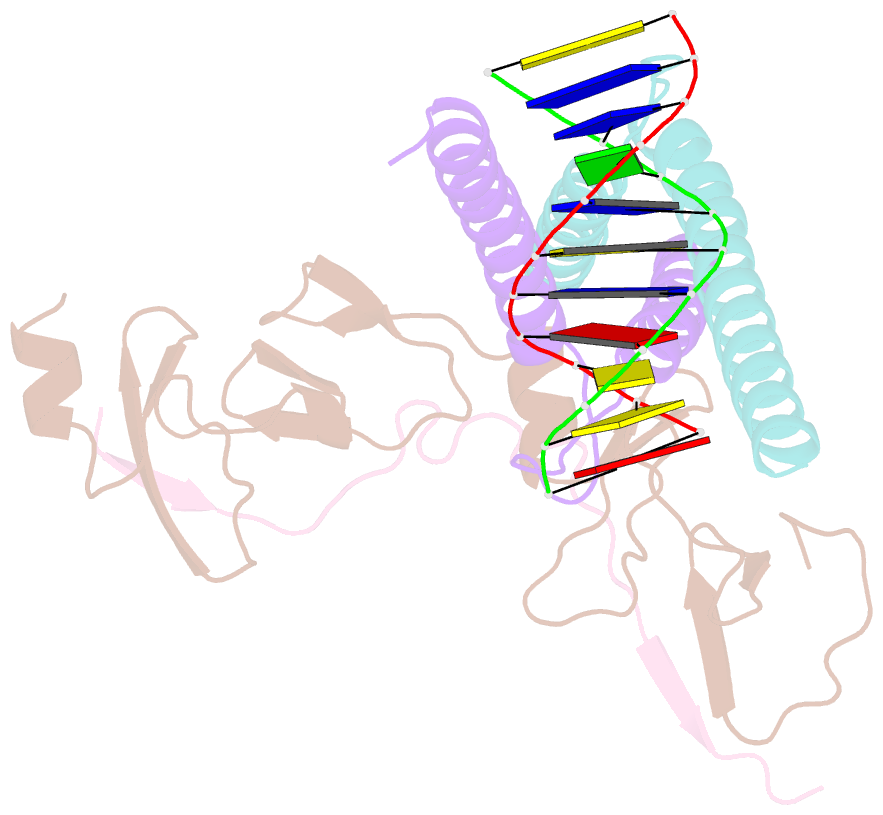
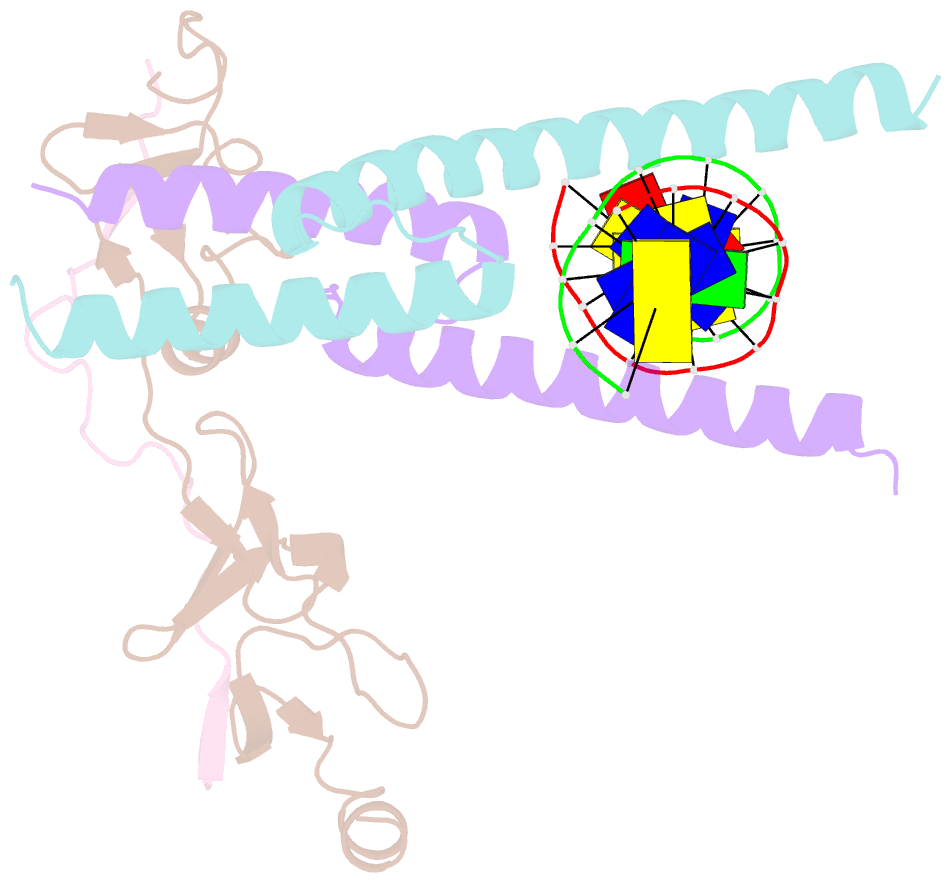
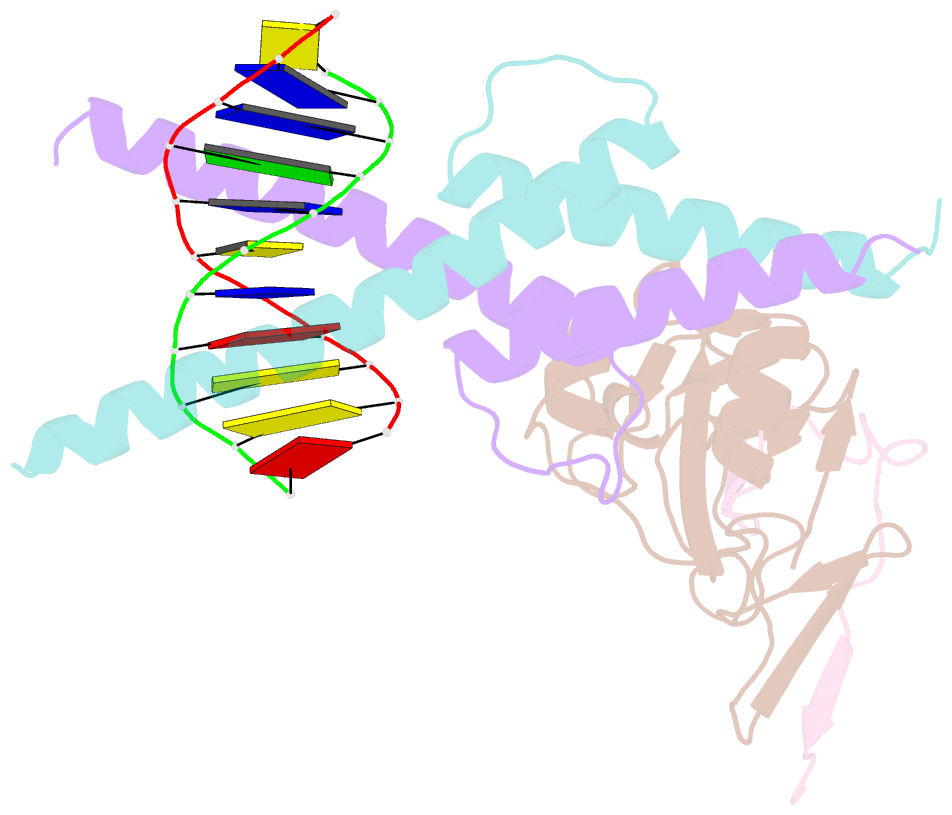
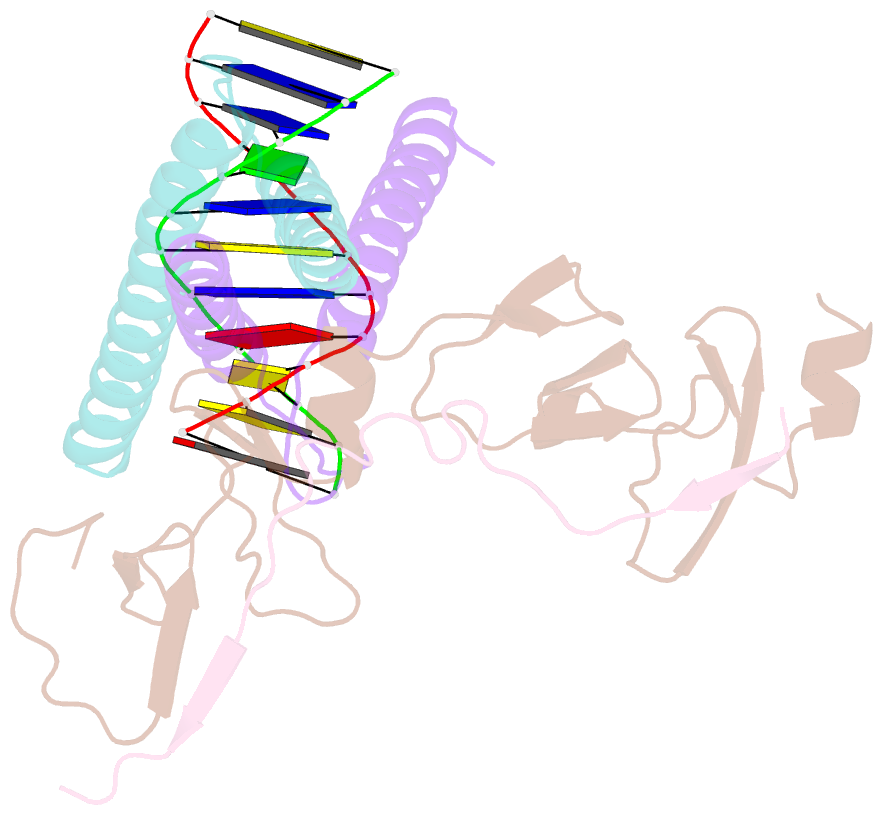
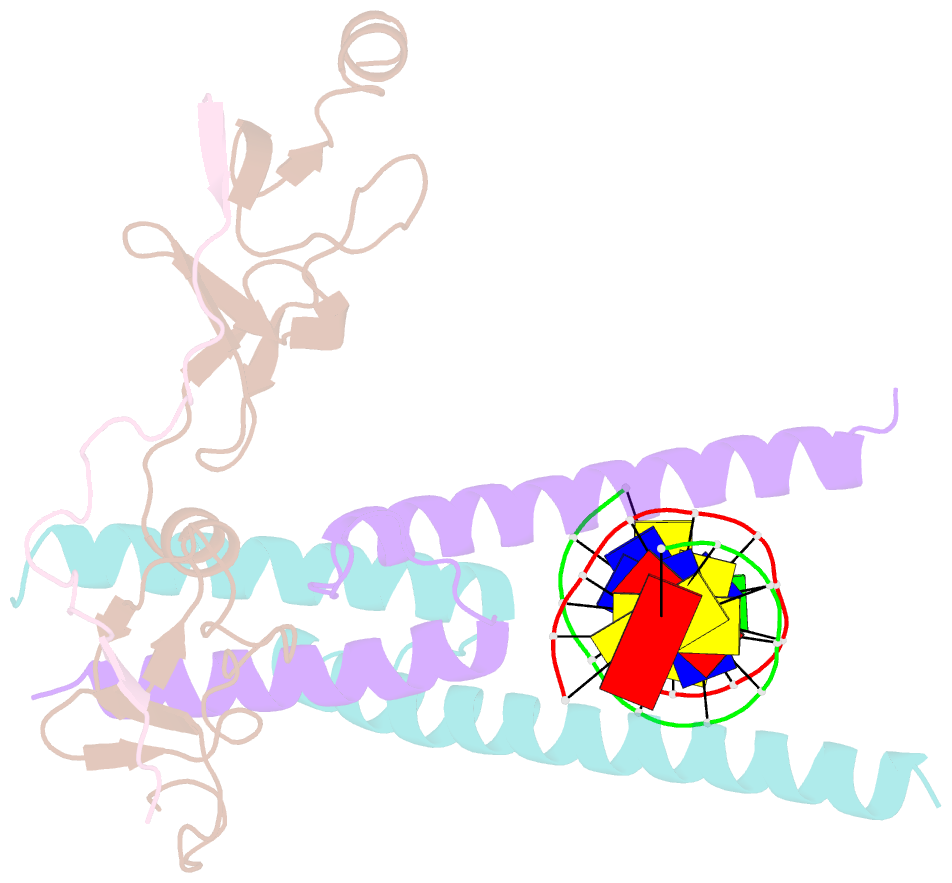
Summary information and primary citation
- PDB-id
- 2ypa; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- immune system
- Method
- X-ray (2.8 Å)
- Summary
- Structure of the scl:e47:lmo2:ldb1 complex bound to DNA
- Reference
- El Omari K, Hoosdally SJ, Tuladhar K, Karia D, Hall-Ponsele E, Platonova O, Vyas P, Patient R, Porcher C, Mancini EJ (2013): "Structural Basis for Lmo2-Driven Recruitment of the Scl:E47bHLH Heterodimer to Hematopoietic-Specific Transcriptional Targets." Cell Rep., 4, 135. doi: 10.1016/J.CELREP.2013.06.008.
- Abstract
- Cell fate is governed by combinatorial actions of transcriptional regulators assembling into multiprotein complexes. However, the molecular details of how these complexes form are poorly understood. One such complex, which contains the basic-helix-loop-helix heterodimer SCL:E47 and bridging proteins LMO2:LDB1, critically regulates hematopoiesis and induces T cell leukemia. Here, we report the crystal structure of (SCL:E47)bHLH:LMO2:LDB1LID bound to DNA, providing a molecular account of the network of interactions assembling this complex. This reveals an unexpected role for LMO2. Upon binding to SCL, LMO2 induces new hydrogen bonds in SCL:E47, thereby strengthening heterodimer formation. This imposes a rotation movement onto E47 that weakens the heterodimer:DNA interaction, shifting the main DNA-binding activity onto additional protein partners. Along with biochemical analyses, this illustrates, at an atomic level, how hematopoietic-specific SCL sequesters ubiquitous E47 and associated cofactors and supports SCL's reported DNA-binding-independent functions. Importantly, this work will drive the design of small molecules inhibiting leukemogenic processes.