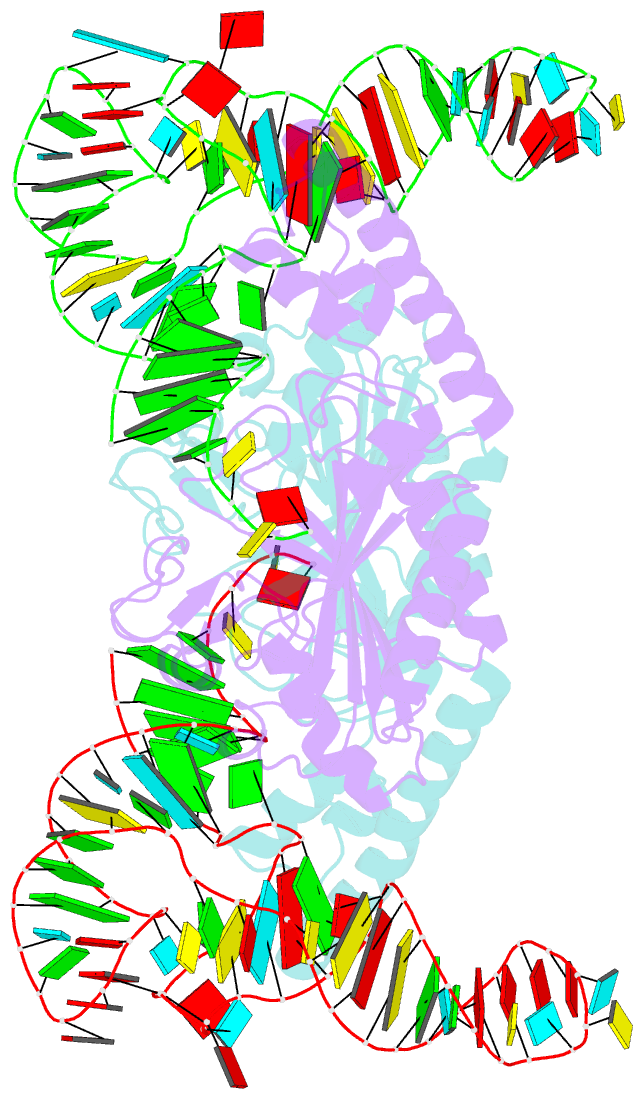
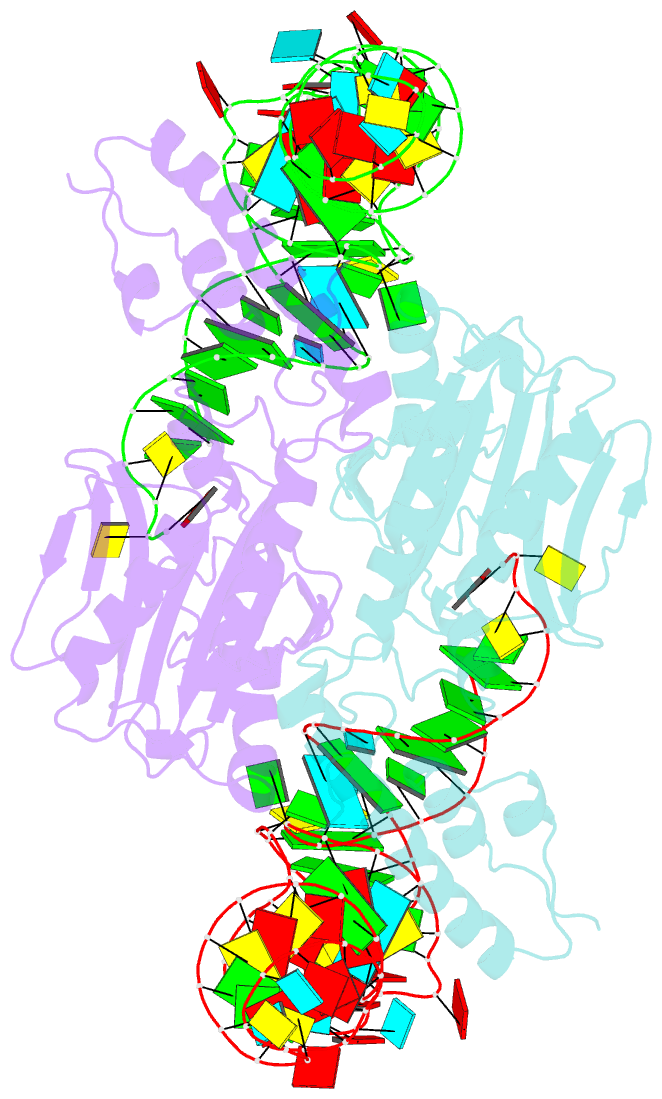
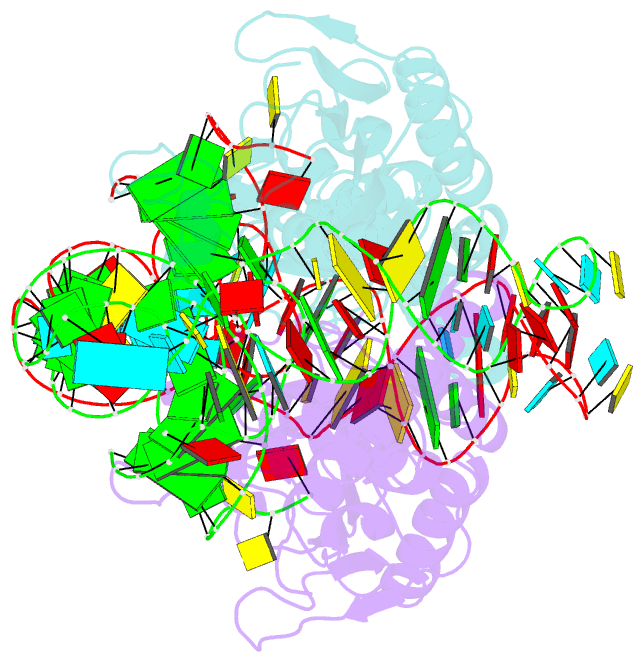
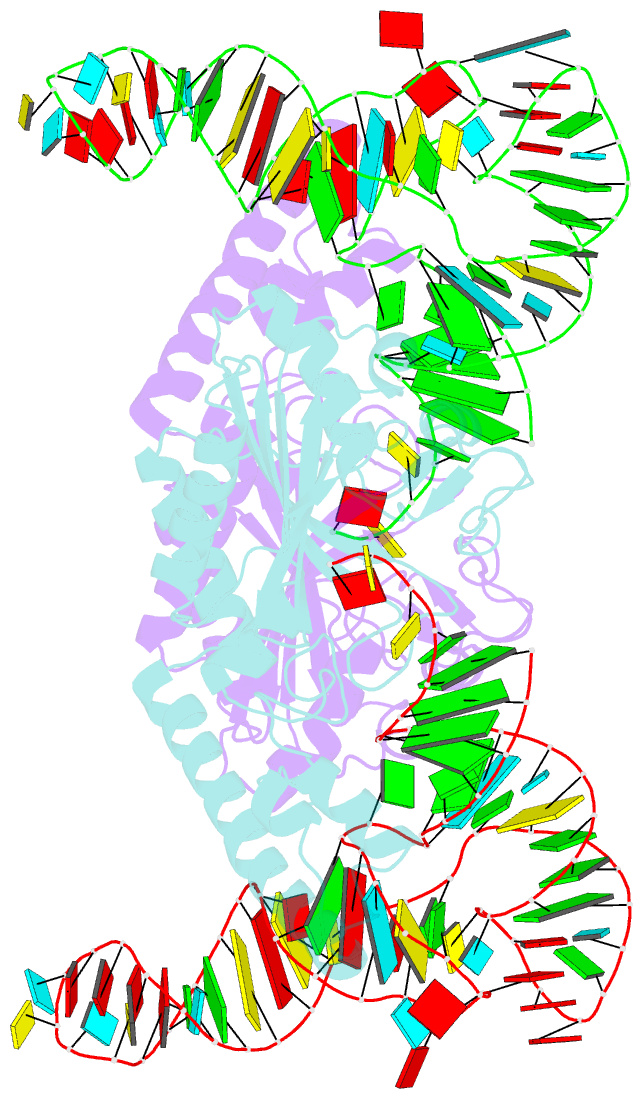
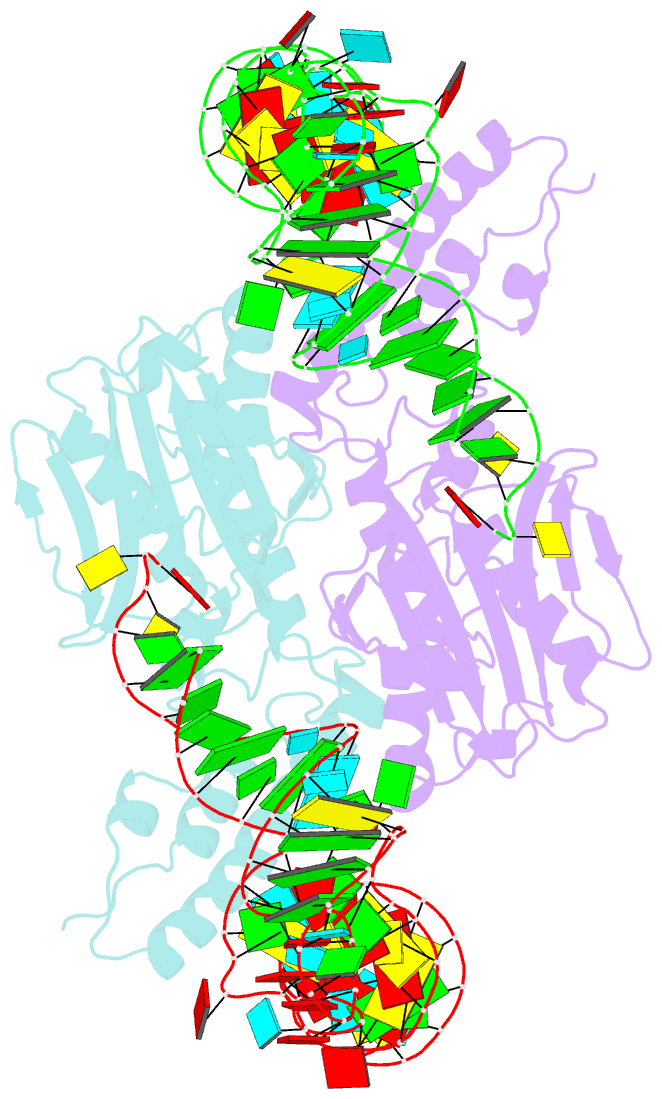
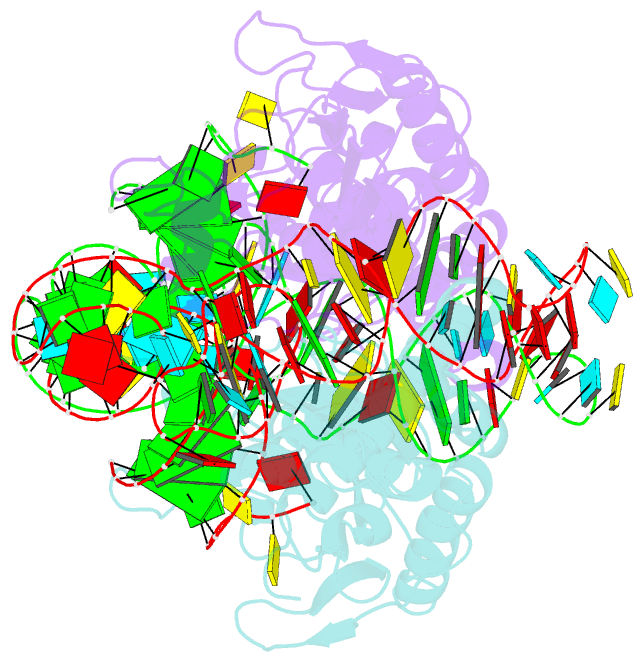
Summary information and primary citation
- PDB-id
- 2zni; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase-RNA
- Method
- X-ray (3.1 Å)
- Summary
- Crystal structure of pyrrolysyl-trna synthetase-trna(pyl) complex from desulfitobacterium hafniense
- Reference
- Nozawa K, O'Donoghue P, Gundllapalli S, Araiso Y, Ishitani R, Umehara T, Soll D, Nureki O (2009): "Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality." Nature, 457, 1163-1167. doi: 10.1038/nature07611.
- Abstract
- Pyrrolysine (Pyl), the 22nd natural amino acid, is genetically encoded by UAG and inserted into proteins by the unique suppressor tRNA(Pyl) (ref. 1). The Methanosarcinaceae produce Pyl and express Pyl-containing methyltransferases that allow growth on methylamines. Homologous methyltransferases and the Pyl biosynthetic and coding machinery are also found in two bacterial species. Pyl coding is maintained by pyrrolysyl-tRNA synthetase (PylRS), which catalyses the formation of Pyl-tRNA(Pyl) (refs 4, 5). Pyl is not a recent addition to the genetic code. PylRS was already present in the last universal common ancestor; it then persisted in organisms that utilize methylamines as energy sources. Recent protein engineering efforts added non-canonical amino acids to the genetic code. This technology relies on the directed evolution of an 'orthogonal' tRNA synthetase-tRNA pair in which an engineered aminoacyl-tRNA synthetase (aaRS) specifically and exclusively acylates the orthogonal tRNA with a non-canonical amino acid. For Pyl the natural evolutionary process developed such a system some 3 billion years ago. When transformed into Escherichia coli, Methanosarcina barkeri PylRS and tRNA(Pyl) function as an orthogonal pair in vivo. Here we show that Desulfitobacterium hafniense PylRS-tRNA(Pyl) is an orthogonal pair in vitro and in vivo, and present the crystal structure of this orthogonal pair. The ancient emergence of PylRS-tRNA(Pyl) allowed the evolution of unique structural features in both the protein and the tRNA. These structural elements manifest an intricate, specialized aaRS-tRNA interaction surface that is highly distinct from those observed in any other known aaRS-tRNA complex; it is this general property that underlies the molecular basis of orthogonality.