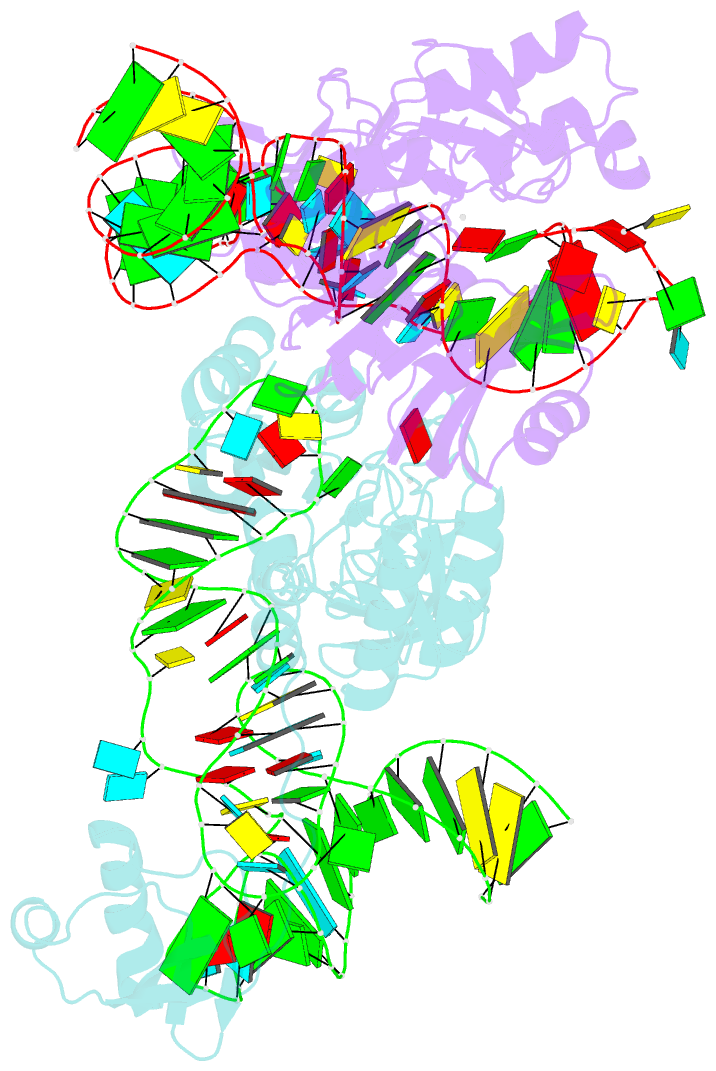
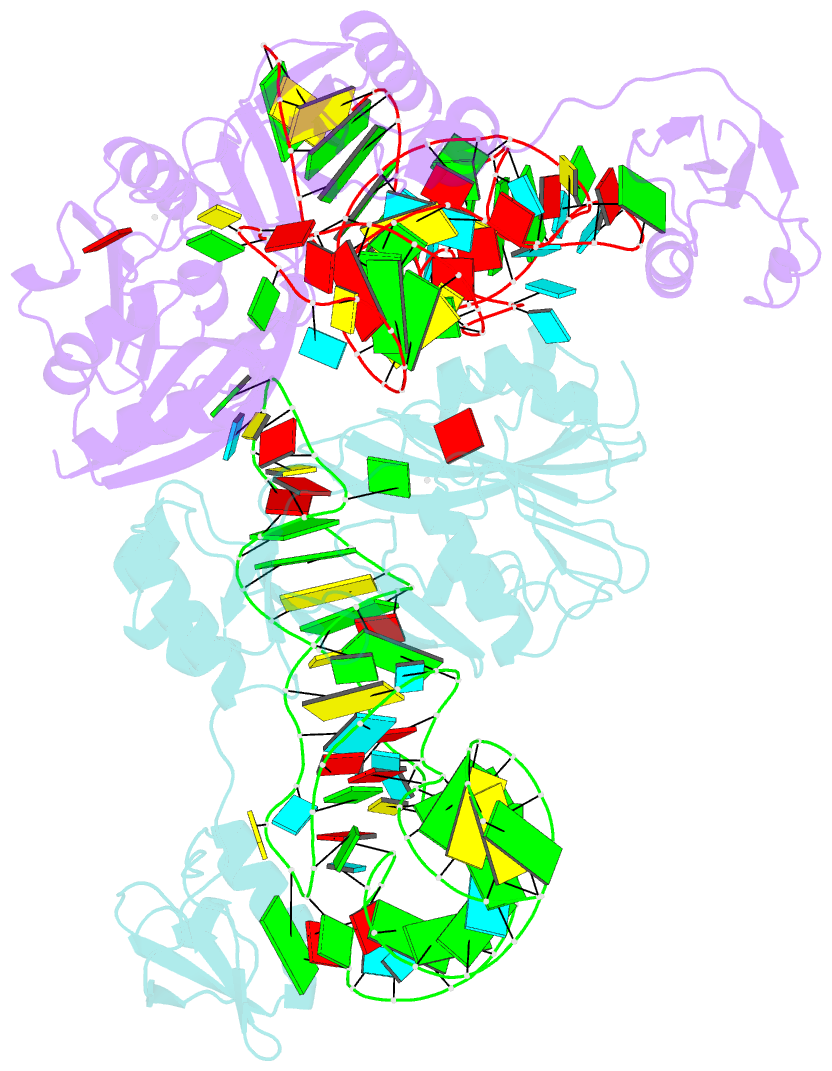
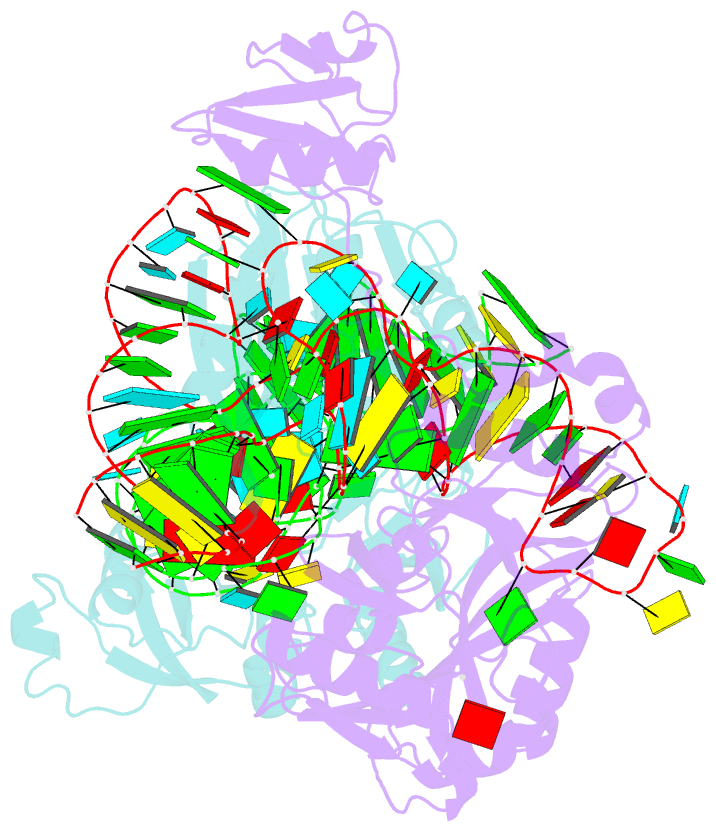
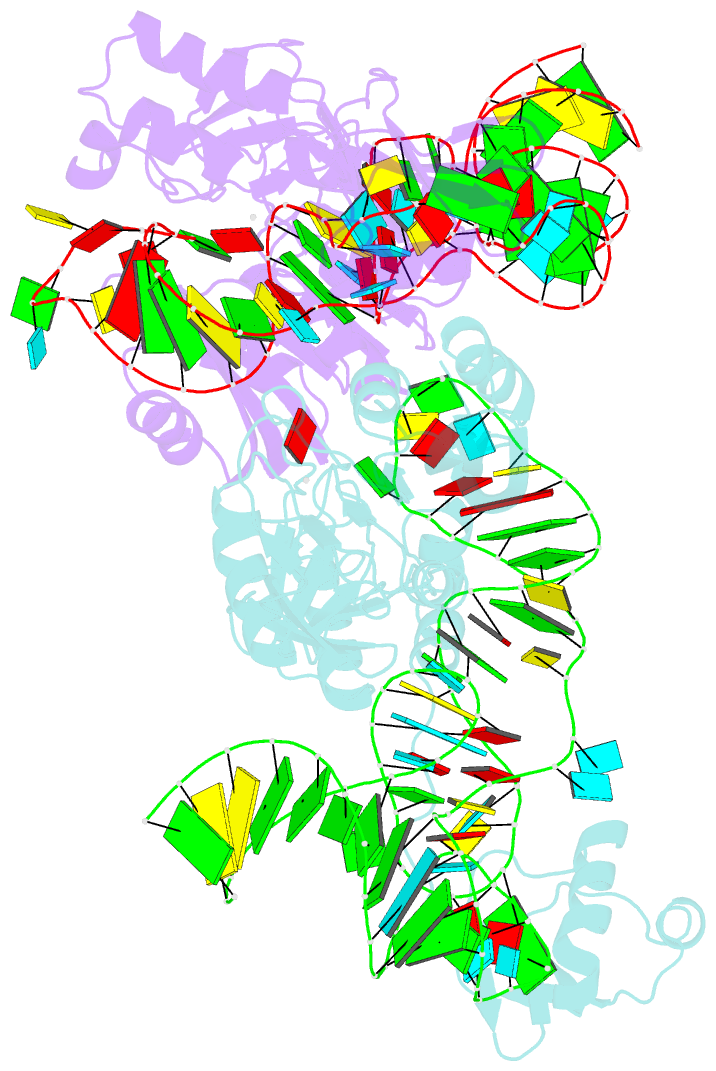
Summary information and primary citation
- PDB-id
-
2zzn;
DSSR-derived features in text and
JSON formats; DNAproDB
- Class
- transferase-RNA
- Method
- X-ray (2.95 Å)
- Summary
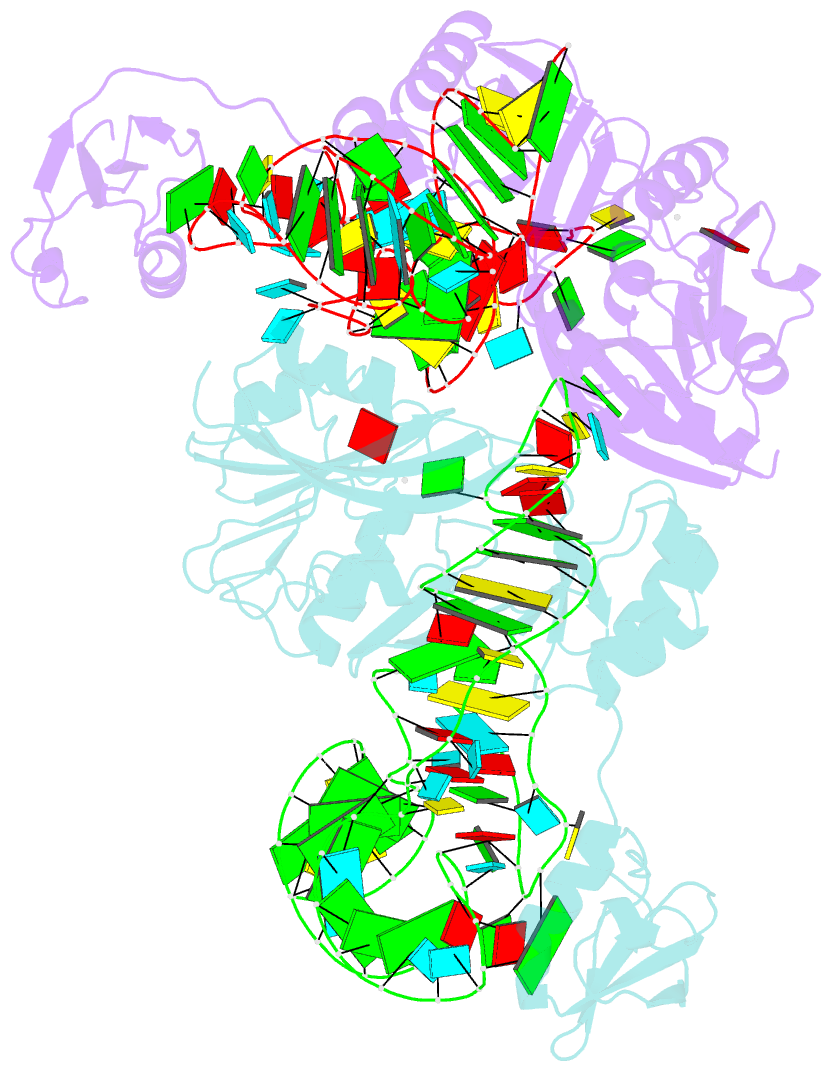
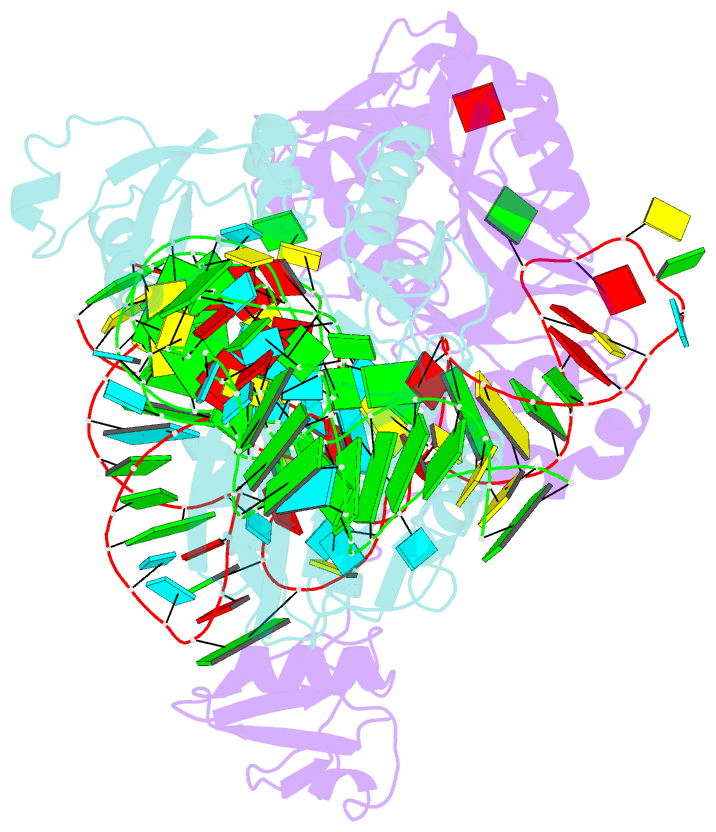
- The complex structure of atrm5 and trnacys
- Reference
-
Goto-Ito S, Ito T, Kuratani M, Bessho Y, Yokoyama S
(2009): "Tertiary
structure checkpoint at anticodon loop modification in
tRNA functional maturation."
Nat.Struct.Mol.Biol., 16,
1109-1115. doi: 10.1038/nsmb.1653.
- Abstract
- tRNA precursors undergo a maturation process, involving
nucleotide modifications and folding into the L-shaped
tertiary structure. The N1-methylguanosine at position 37
(m1G37), 3' adjacent to the anticodon, is essential for
translational fidelity and efficiency. In archaea and
eukaryotes, Trm5 introduces the m1G37 modification into all
tRNAs bearing G37. Here we report the crystal structures of
archaeal Trm5 (aTrm5) in complex with tRNA(Leu) or
tRNA(Cys). The D2-D3 domains of aTrm5 discover and modify
G37, independently of the tRNA sequences. D1 is connected
to D2-D3 through a flexible linker and is designed to
recognize the shape of the tRNA outer corner, as a hallmark
of the completed L shape formation. This interaction by D1
lowers the K(m) value for tRNA, enabling the D2-D3
catalysis. Thus, we propose that aTrm5 provides the
tertiary structure checkpoint in tRNA maturation.