Summary information and primary citation
- PDB-id
- 3adi; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- gene regulation-RNA
- Method
- X-ray (3.2 Å)
- Summary
- Structure of arabidopsis hyl1 and its molecular implications for mirna processing
- Reference
- Yang SW, Chen HY, Yang J, Machida S, Chua NH, Yuan YA (2010): "Structure of arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing." Structure, 18, 594-605. doi: 10.1016/j.str.2010.02.006.
- Abstract
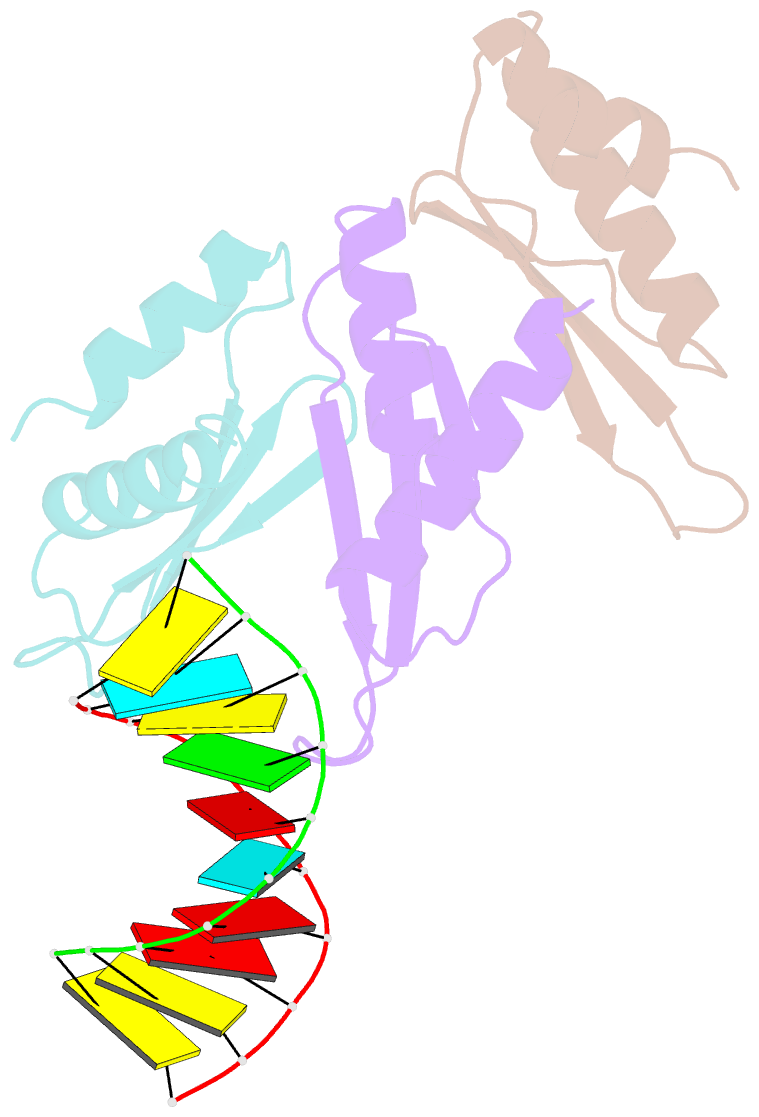
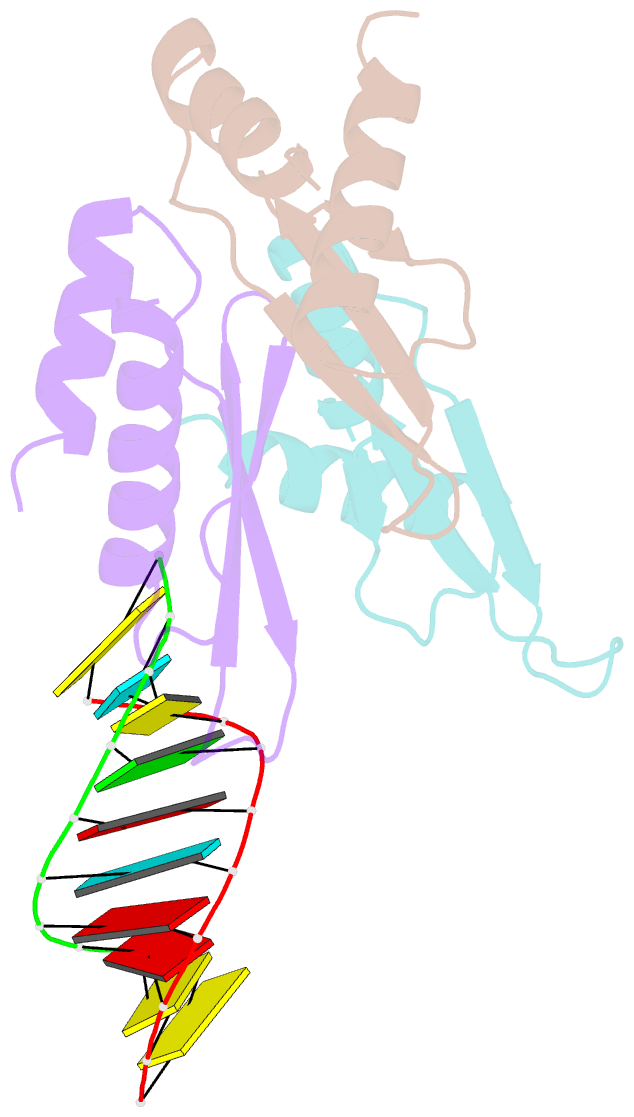
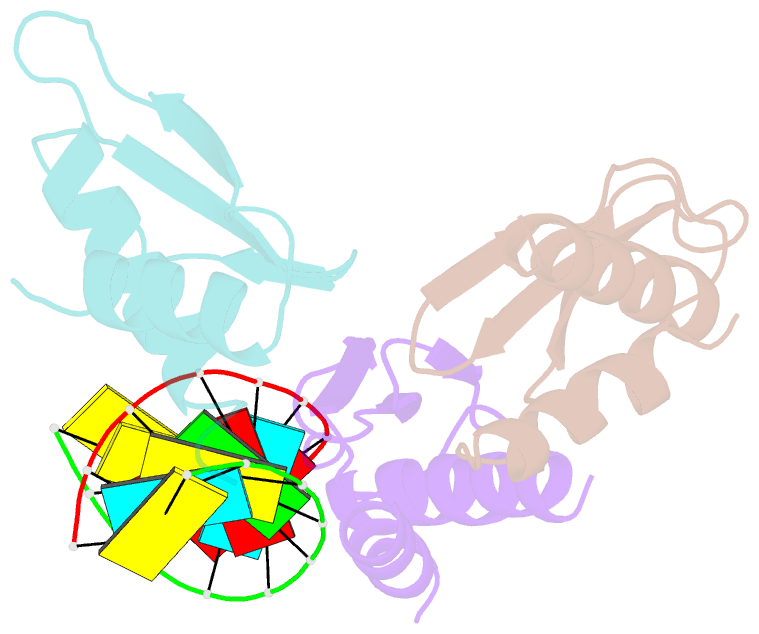
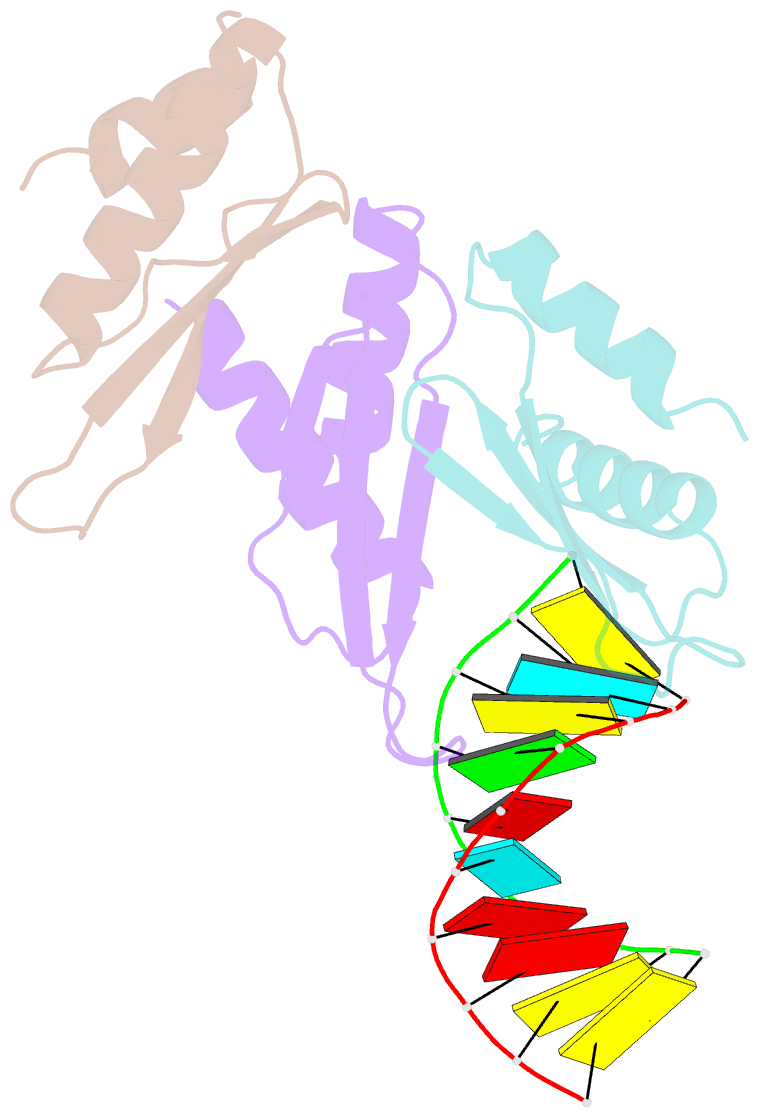
- The Arabidopsis HYPONASTIC LEAVES1 (HYL1) is a double-stranded RNA-binding protein that forms a complex with DICER-LIKE1 (DCL1) and SERRATE to facilitate processing of primary miRNAs into microRNAs (miRNAs). However, the structural mechanisms of miRNA maturation by this complex are poorly understood. Here, we present the crystal structures of double-stranded RNA binding domains (dsRBD1 and dsRBD2) of HYL1 and HYL1 dsRBD1 (HR1)/dsRNA complex as well as human TRBP2 dsRBD2 (TR2)/dsRNA complex for comparison analysis. Structural and functional study demonstrates that both HR1 and TR2 are canonical dsRBDs for dsRNA binding, whereas HR2 of HYL1 is a non-canonical dsRBD harboring a putative dimerization interface. Domain swapping within the context of HYL1 demonstrates that TR2 can supplant the function of HR1 in vitro and in vivo. Further biochemical analyses suggest that HYL1 probably binds to the miRNA/miRNA( *) region of precursors as a dimer mediated by HR2.