Summary information and primary citation
- PDB-id
- 3avt; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- translation, transferase-RNA
- Method
- X-ray (2.607 Å)
- Summary
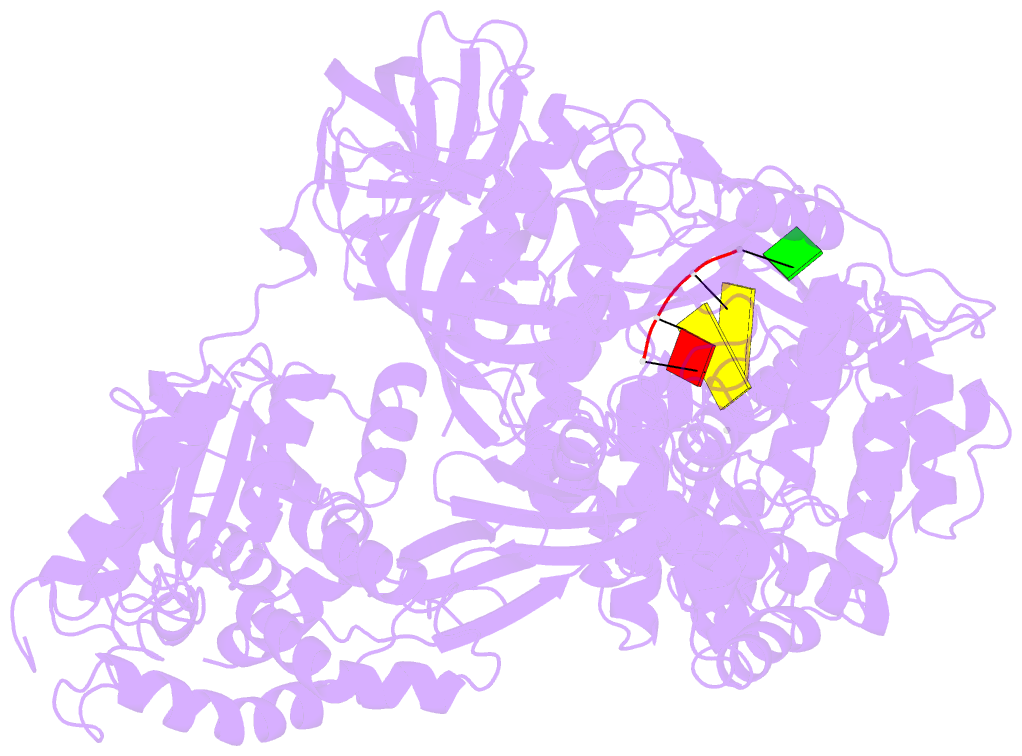
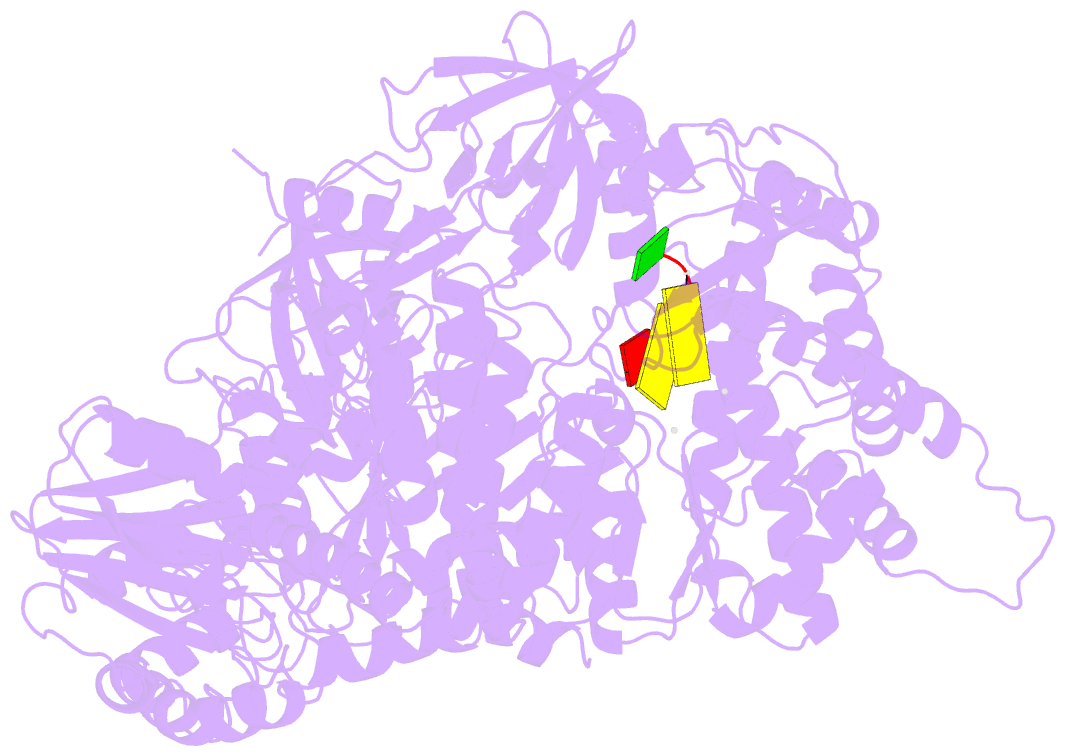
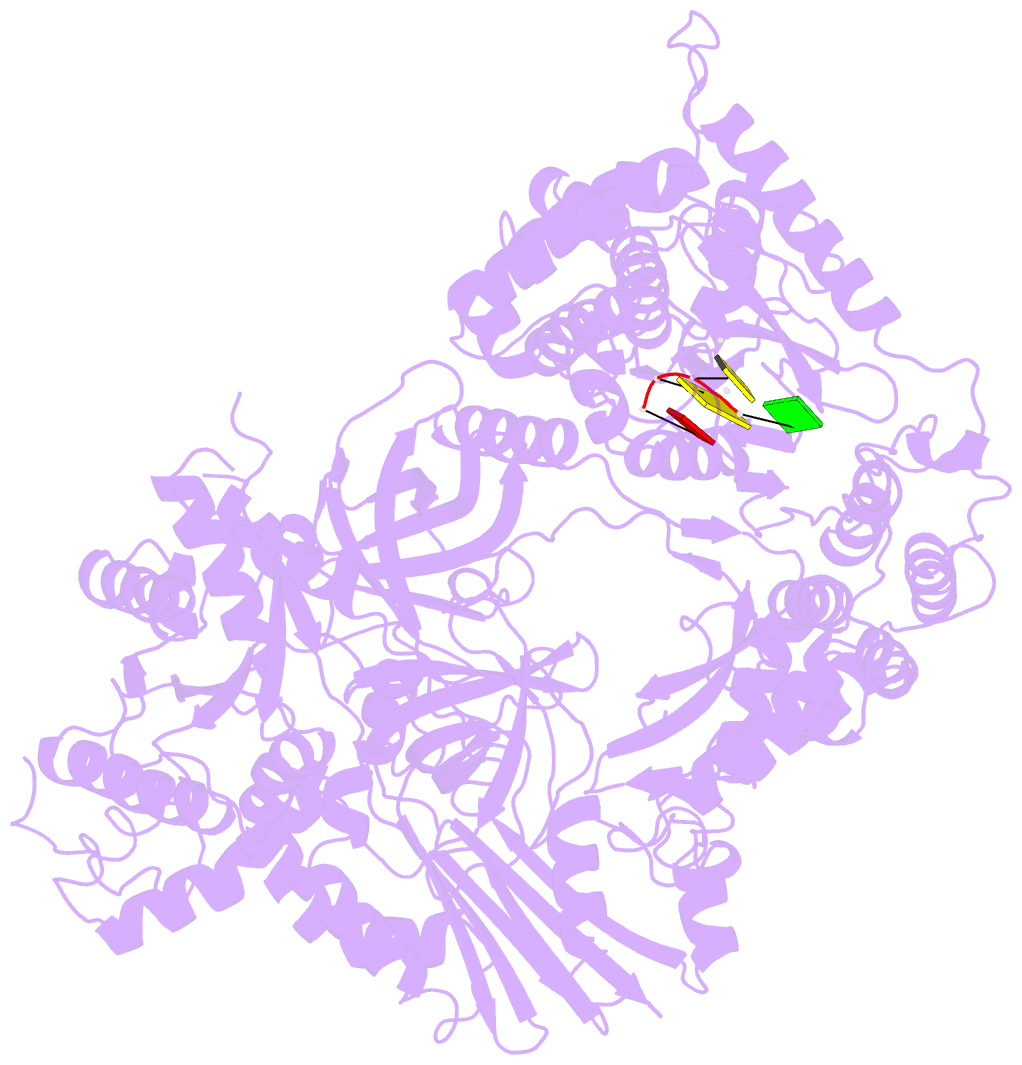
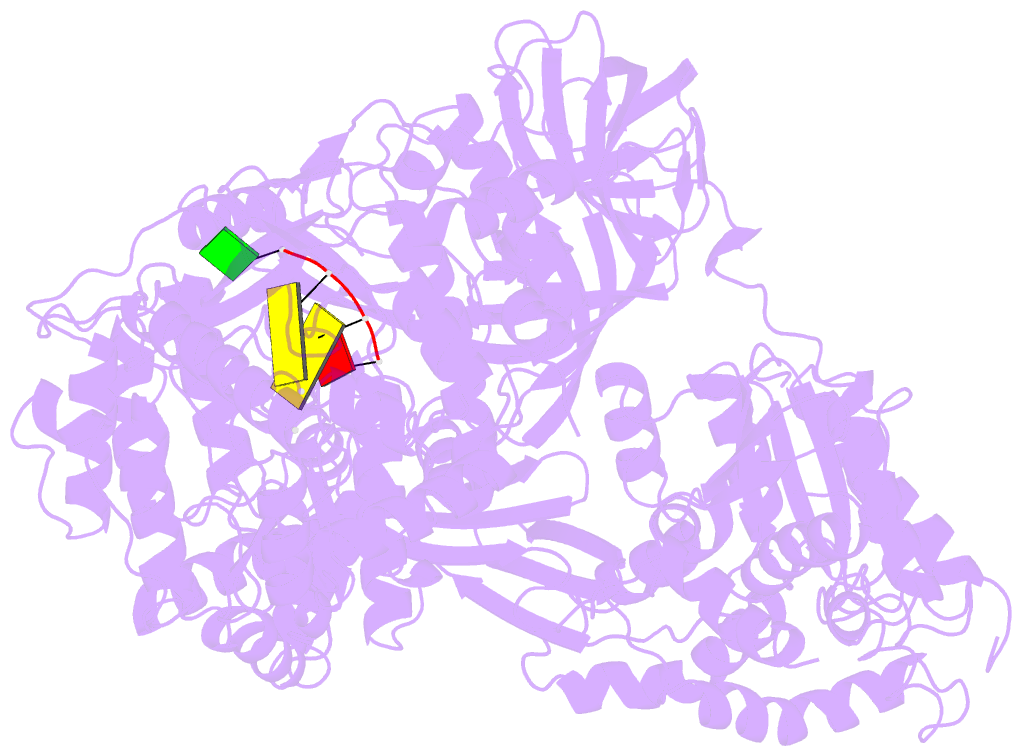
- Structure of viral RNA polymerase complex 1
- Reference
- Takeshita D, Tomita K (2012): "Molecular basis for RNA polymerization by Q beta replicase." Nat.Struct.Mol.Biol., 19, 229-237. doi: 10.1038/nsmb.2204.
- Abstract
- Core Qβ replicase comprises the Qβ virus-encoded RNA-dependent RNA polymerase (β-subunit) and the host Escherichia coli translational elongation factors EF-Tu and EF-Ts. The functions of the host proteins in the viral replicase are not clear. Structural analyses of RNA polymerization by core Qβ replicase reveal that at the initiation stage, the 3'-adenine of the template RNA provides a stable platform for de novo initiation. EF-Tu in Qβ replicase forms a template exit channel with the β-subunit. At the elongation stages, the C-terminal region of the β-subunit, assisted by EF-Tu, splits the temporarily double-stranded RNA between the template and nascent RNAs before translocation of the single-stranded template RNA into the exit channel. Therefore, EF-Tu in Qβ replicase modulates RNA elongation processes in a distinct manner from its established function in protein synthesis.