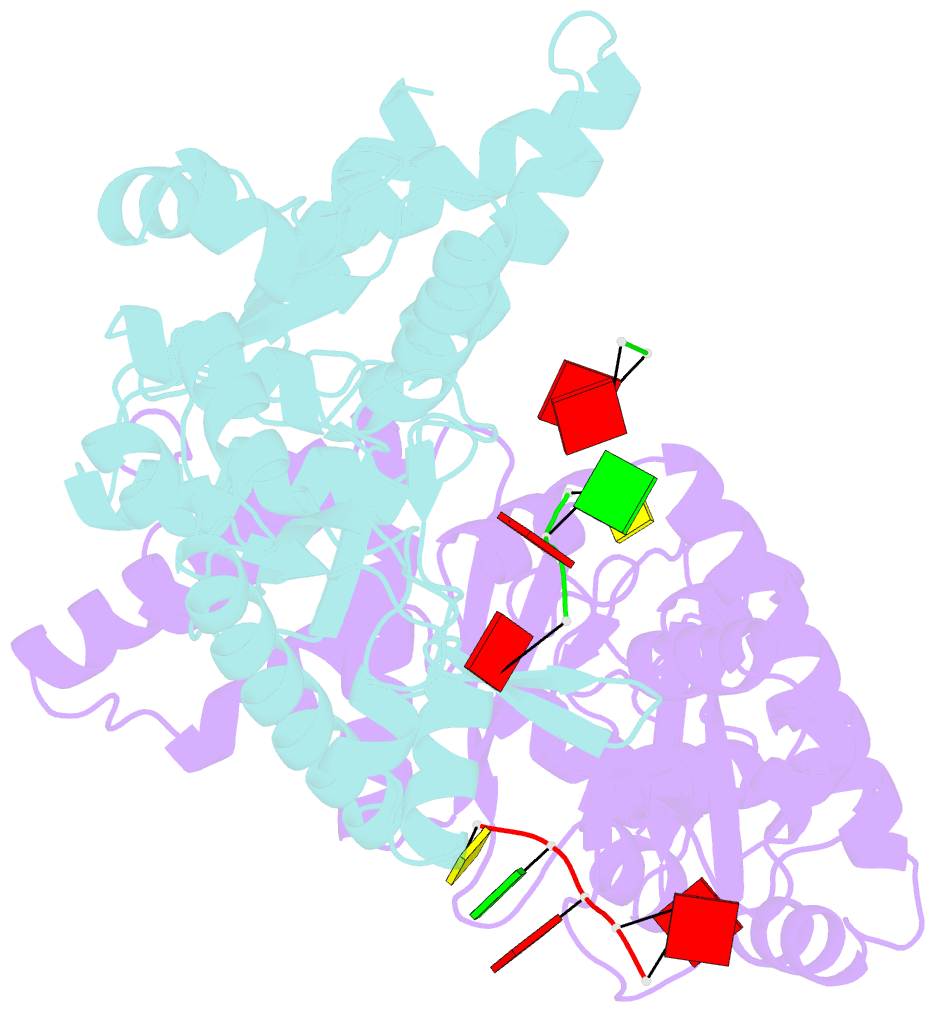
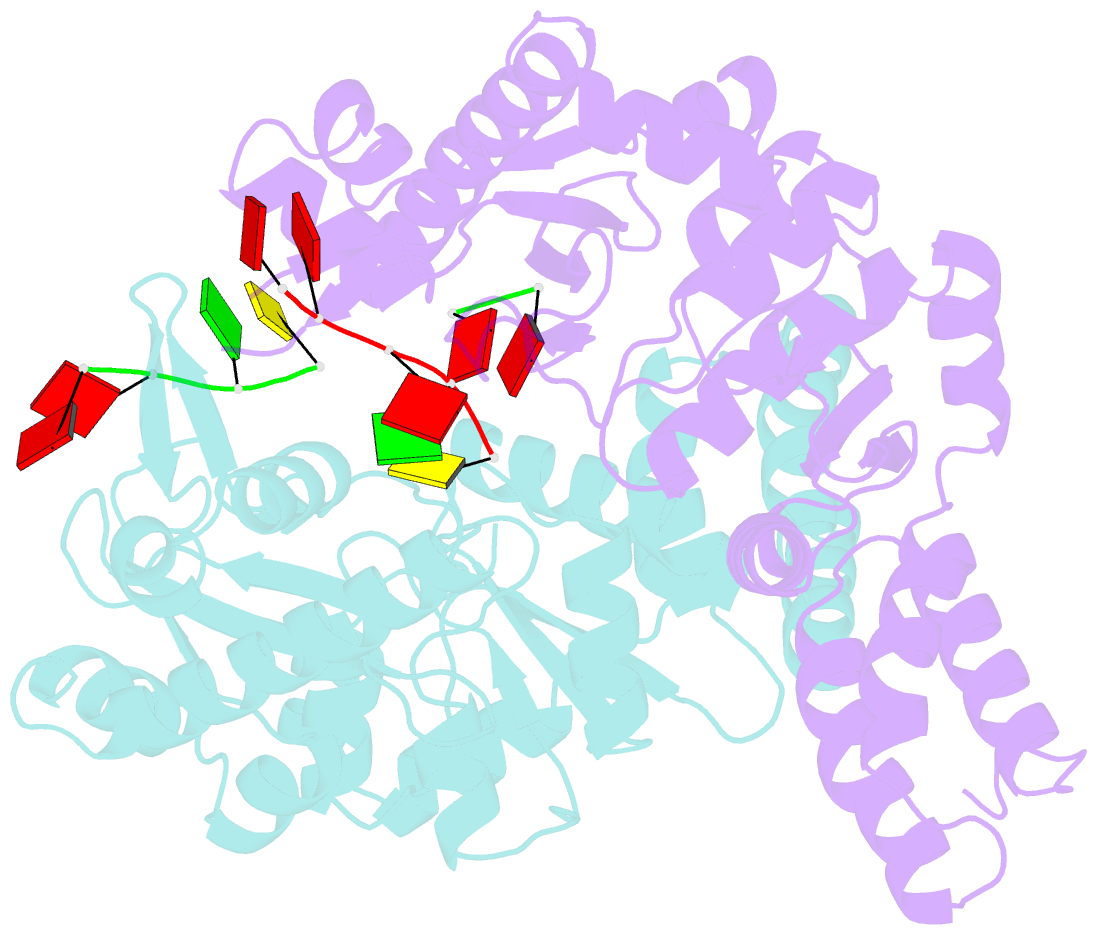
Summary information and primary citation
- PDB-id
- 3b39; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.35 Å)
- Summary
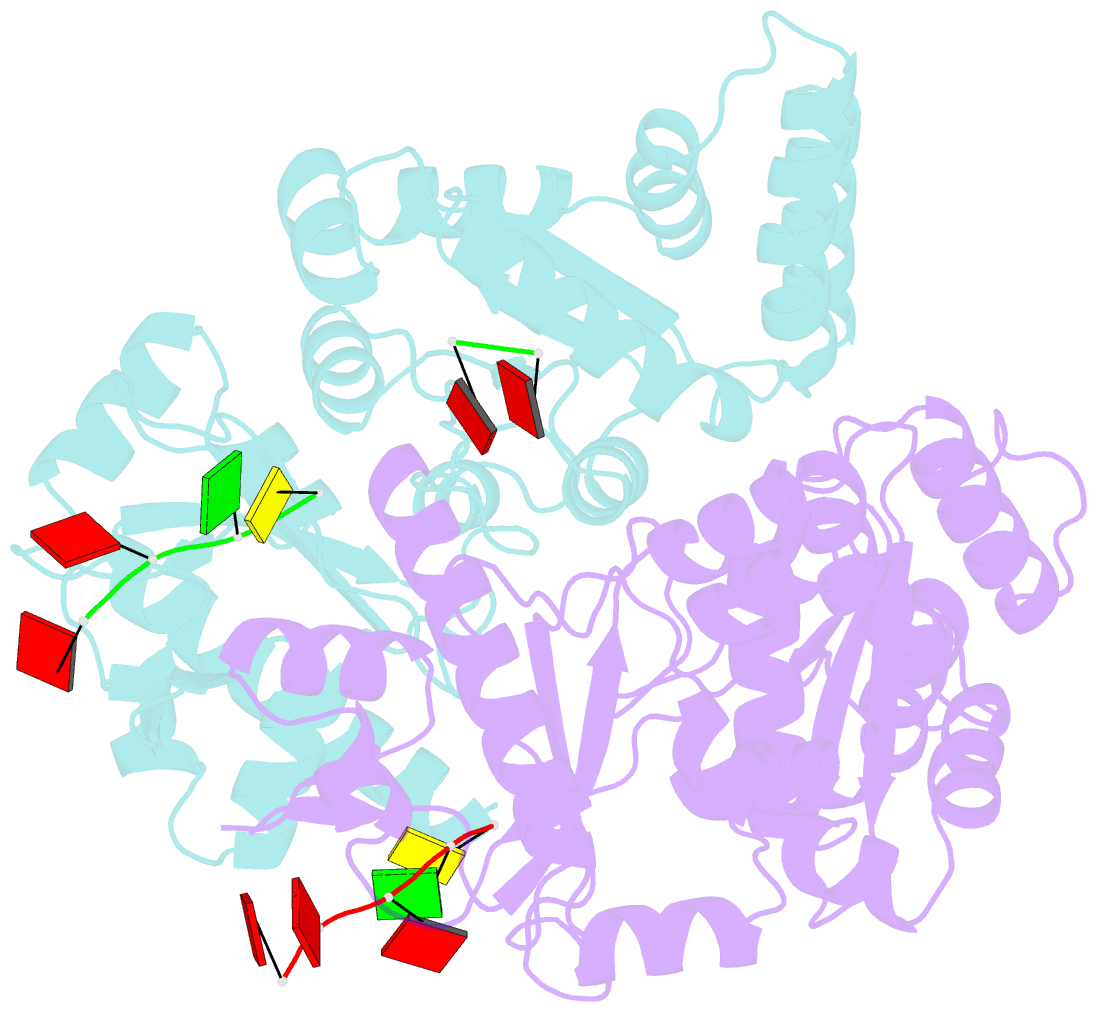
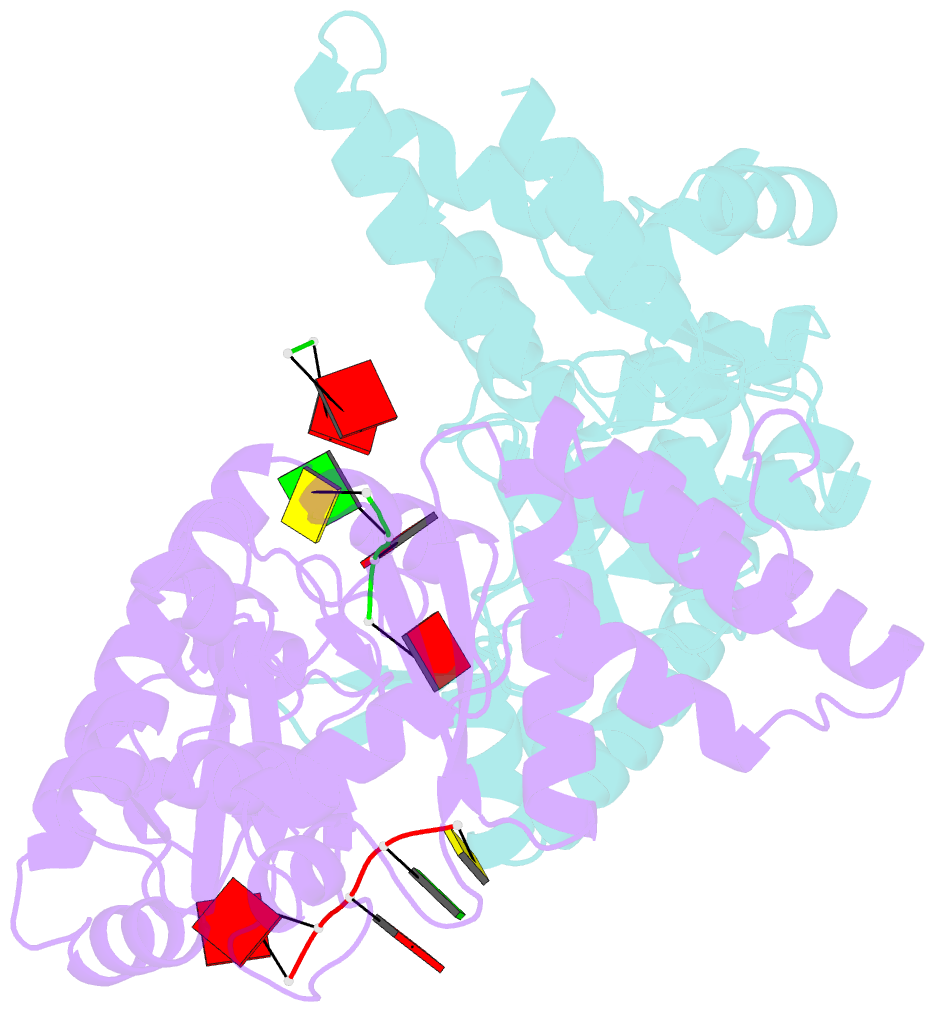
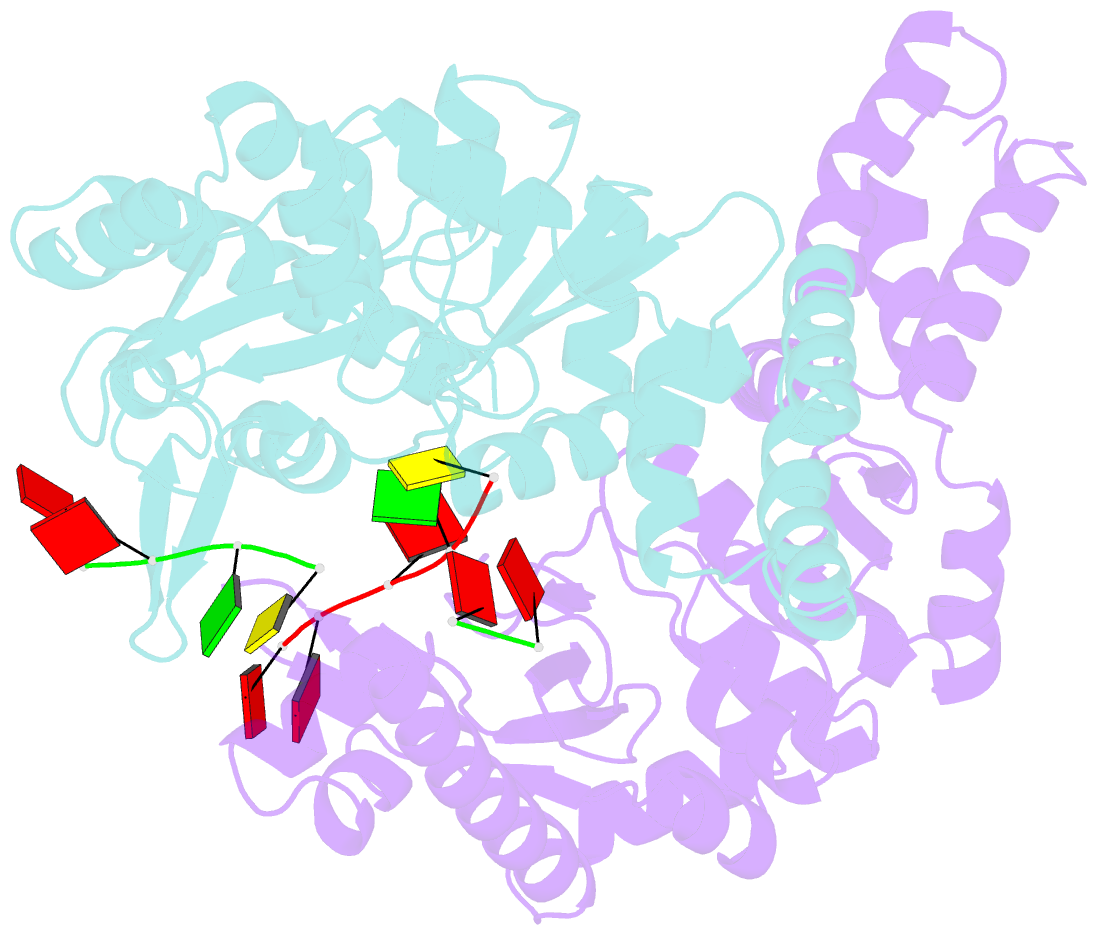
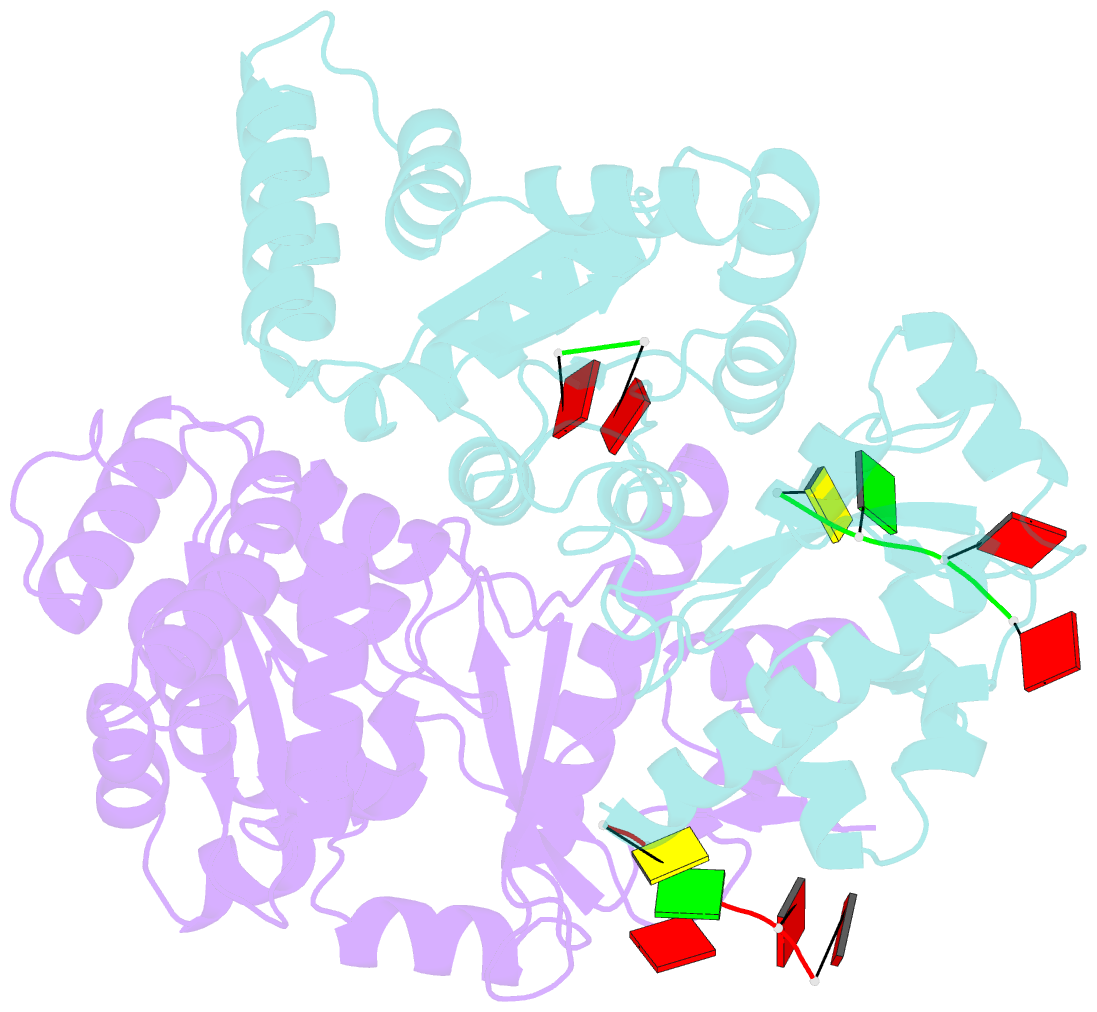
- Structure of the dnag primase catalytic domain bound to ssDNA
- Reference
- Corn JE, Pelton JG, Berger JM (2008): "Identification of a DNA primase template tracking site redefines the geometry of primer synthesis." Nat.Struct.Mol.Biol., 15, 163-169. doi: 10.1038/nsmb.1373.
- Abstract
- Primases are essential RNA polymerases required for the initiation of DNA replication, lagging strand synthesis and replication restart. Many aspects of primase function remain unclear, including how the enzyme associates with a moving nucleic acid strand emanating from a helicase and orients primers for handoff to replisomal components. Using a new screening method to trap transient macromolecular interactions, we determined the structure of the Escherichia coli DnaG primase catalytic domain bound to single-stranded DNA. The structure reveals an unanticipated binding site that engages nucleic acid in two distinct configurations, indicating that it serves as a nonspecific capture and tracking locus for template DNA. Bioinformatic and biochemical analyses show that this evolutionarily constrained region enforces template polarity near the active site and is required for primase function. Together, our findings reverse previous proposals for primer-template orientation and reconcile disparate studies to re-evaluate replication fork organization.