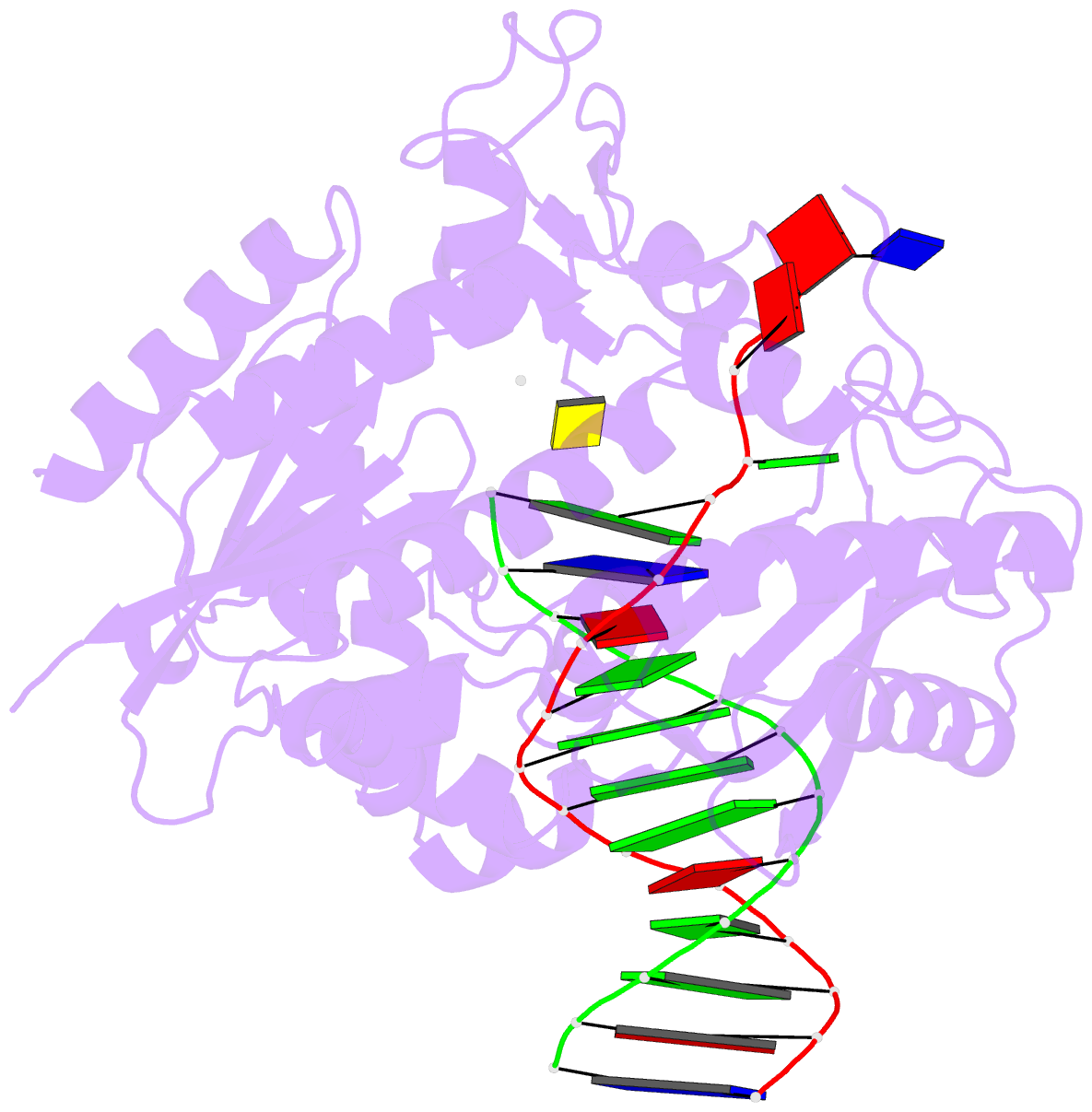
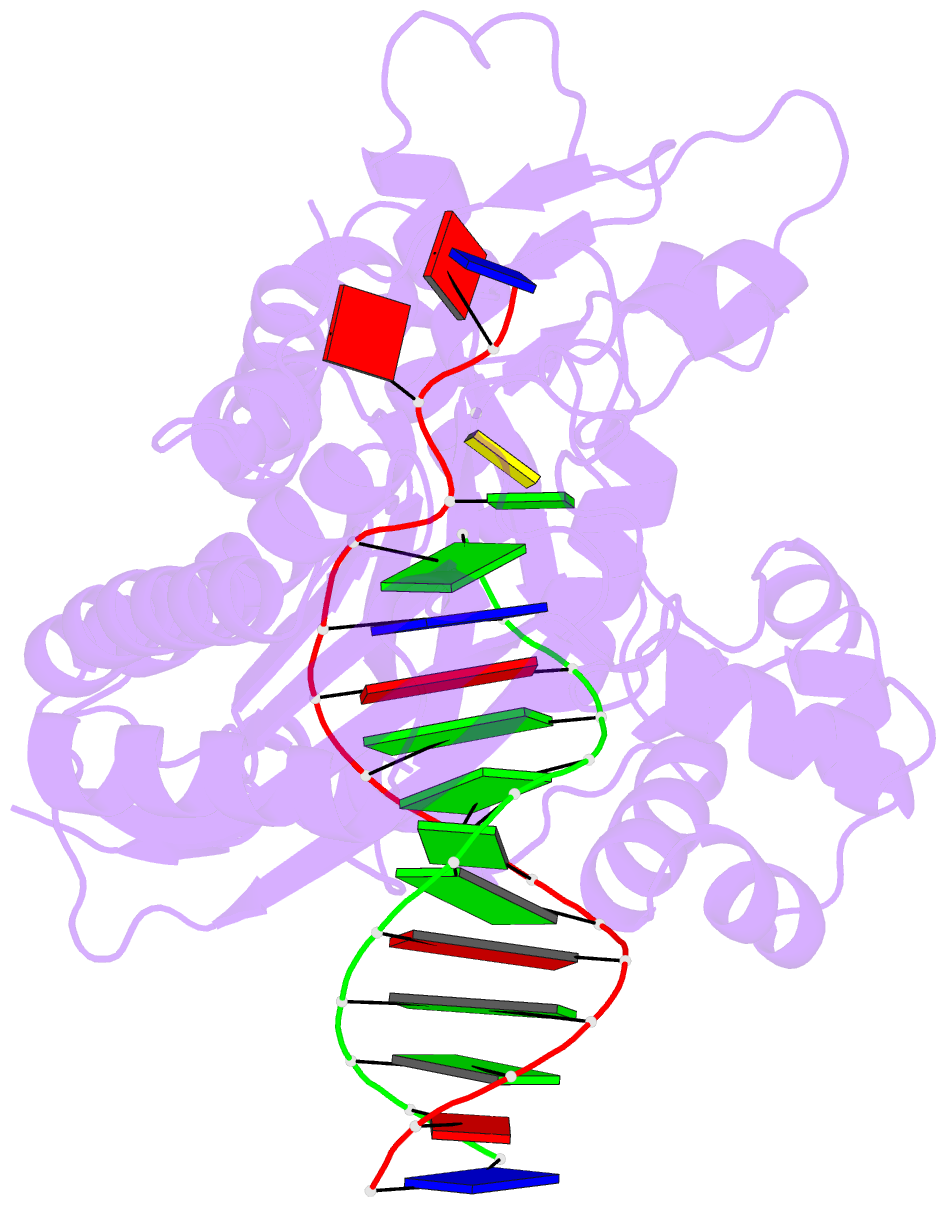
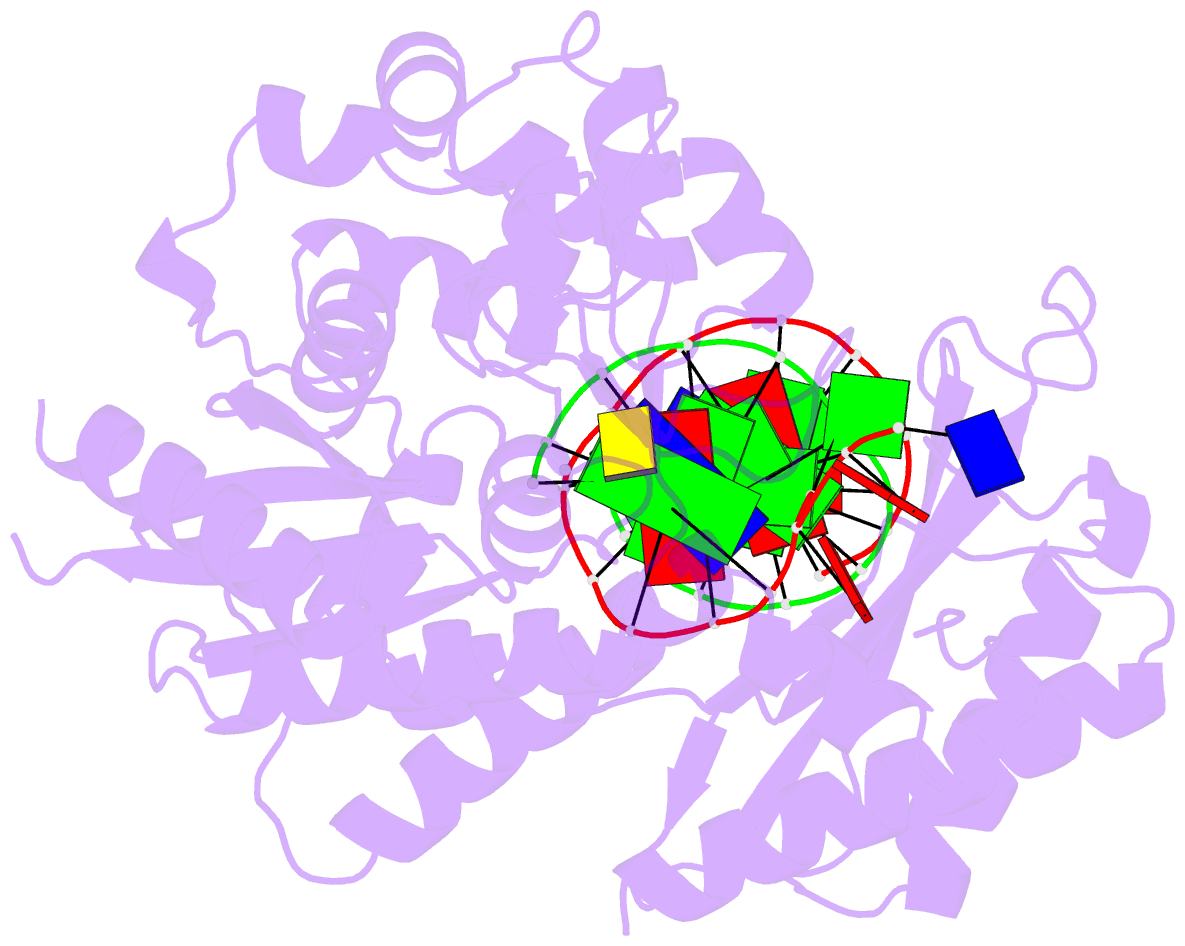
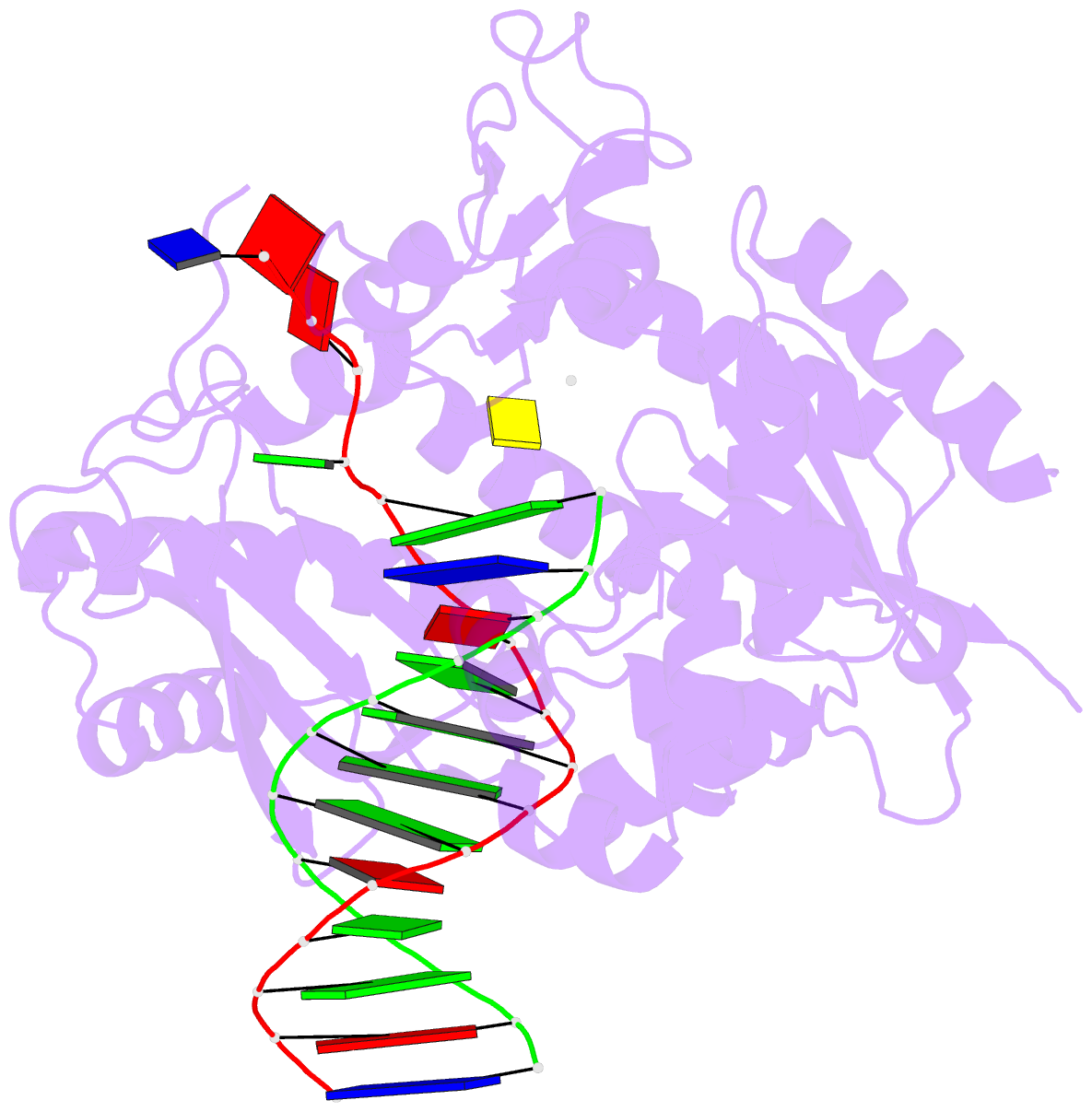
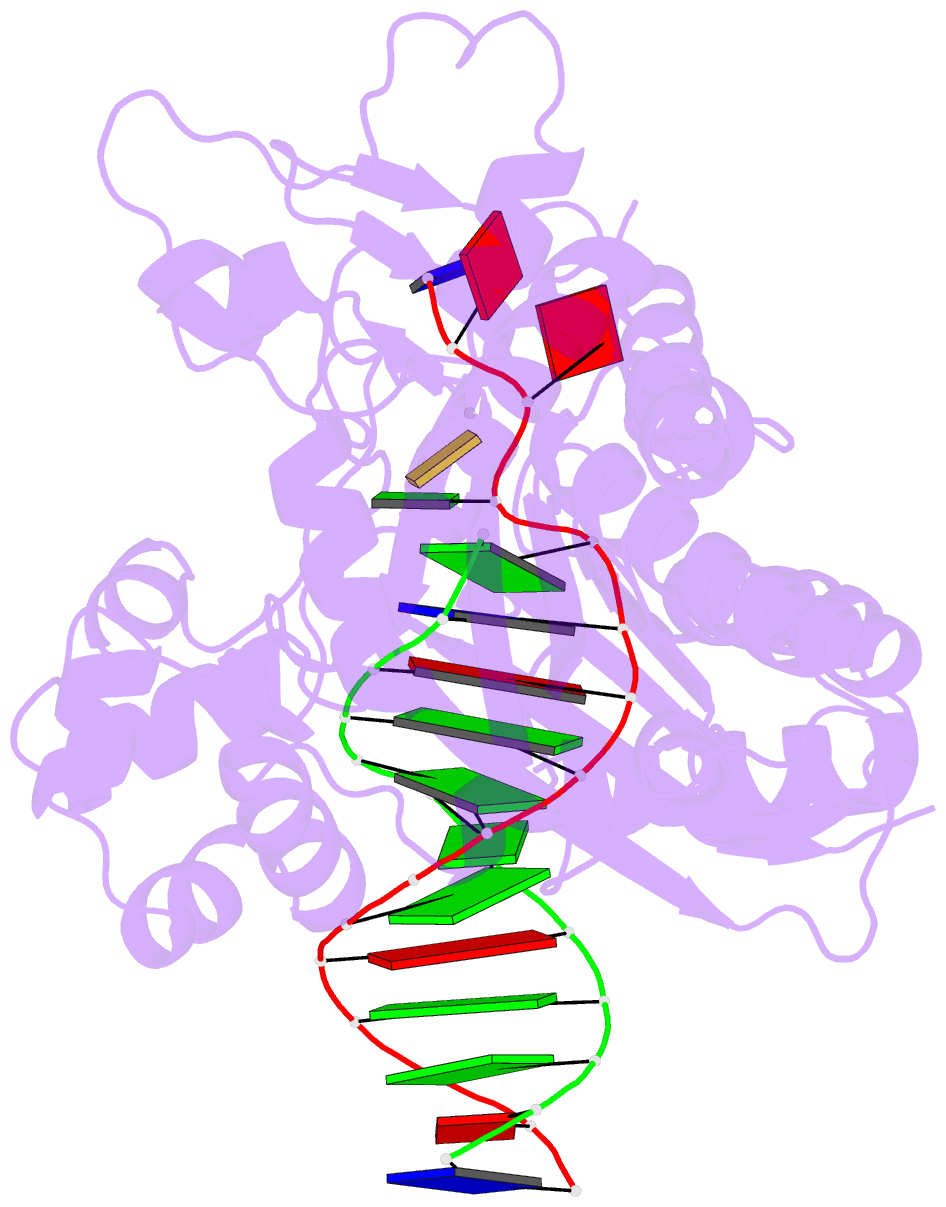
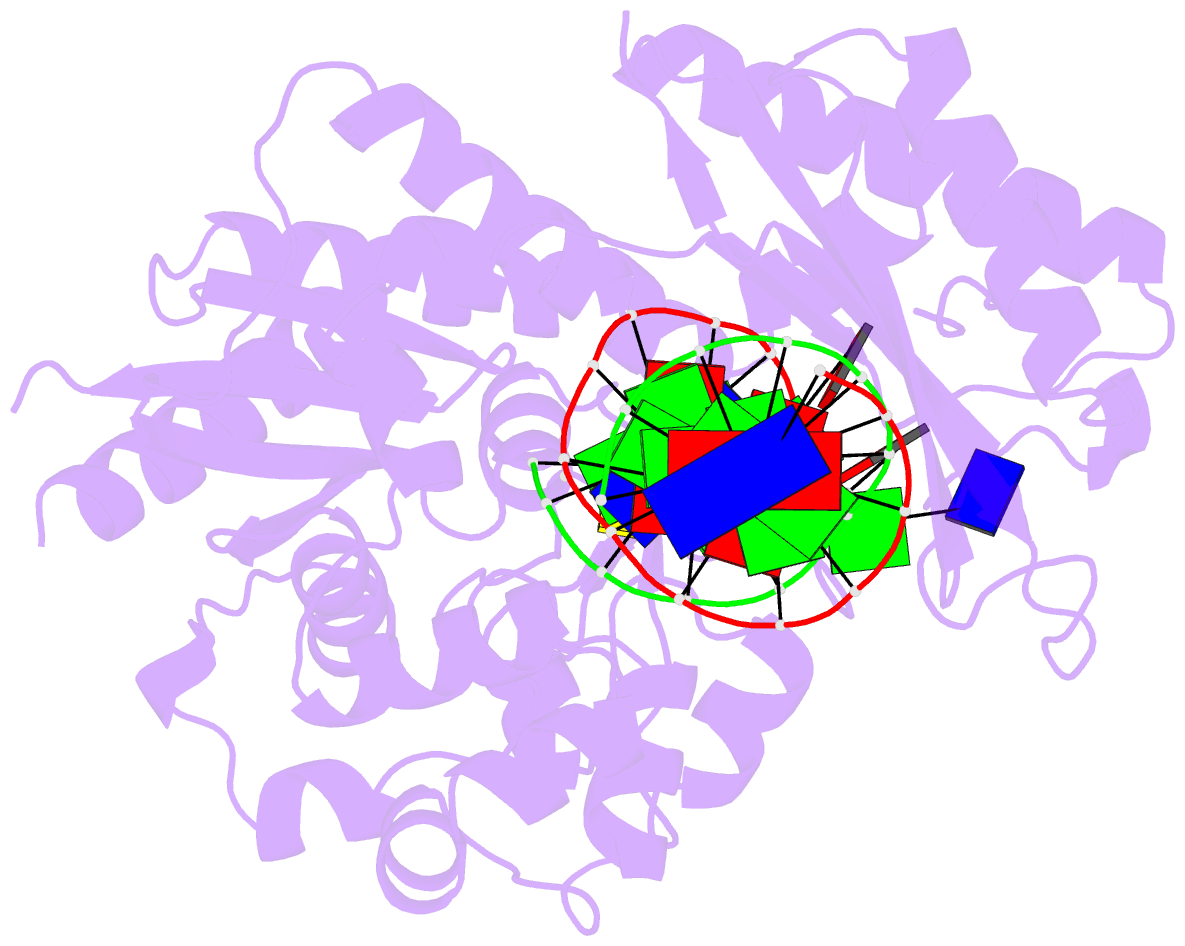
Summary information and primary citation
- PDB-id
- 3bjy; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.41 Å)
- Summary
- Catalytic core of rev1 in complex with DNA (modified template guanine) and incoming nucleotide
- Reference
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK (2008): "Protein-template-directed synthesis across an acrolein-derived DNA adduct by yeast Rev1 DNA polymerase." Structure, 16, 239-245. doi: 10.1016/j.str.2007.12.009.
- Abstract
- Acrolein is generated as the end product of lipid peroxidation and is also a ubiquitous environmental pollutant. Its reaction with the N2 of guanine leads to a cyclic gamma-HOPdG adduct that presents a block to normal replication. We show here that yeast Rev1 incorporates the correct nucleotide C opposite a permanently ring-closed form of gamma-HOPdG (PdG) with nearly the same efficiency as opposite an undamaged G. The structural basis of this action lies in the eviction of the PdG adduct from the Rev1 active site, and the pairing of incoming dCTP with a "surrogate" arginine residue. We also show that yeast Polzeta can carry out the subsequent extension reaction. Together, our studies reveal how the exocyclic PdG adduct is accommodated in a DNA polymerase active site, and they show that the combined action of Rev1 and Polzeta provides for accurate and efficient synthesis through this potentially carcinogenic DNA lesion.