Summary information and primary citation
- PDB-id
- 3brd; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.21 Å)
- Summary
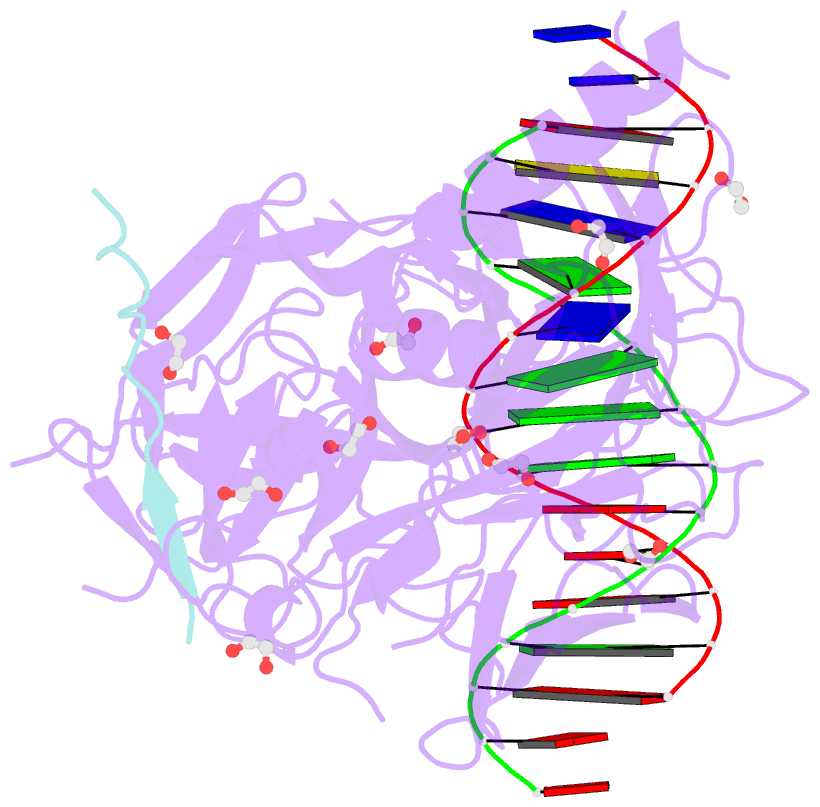
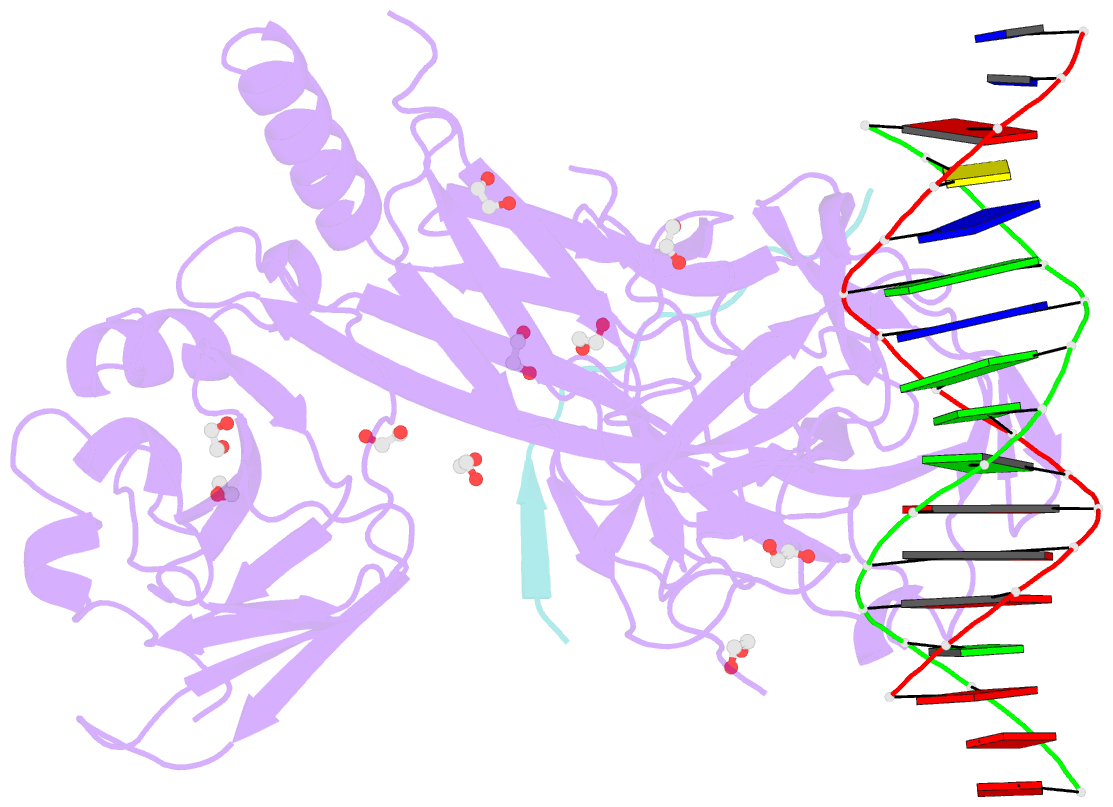
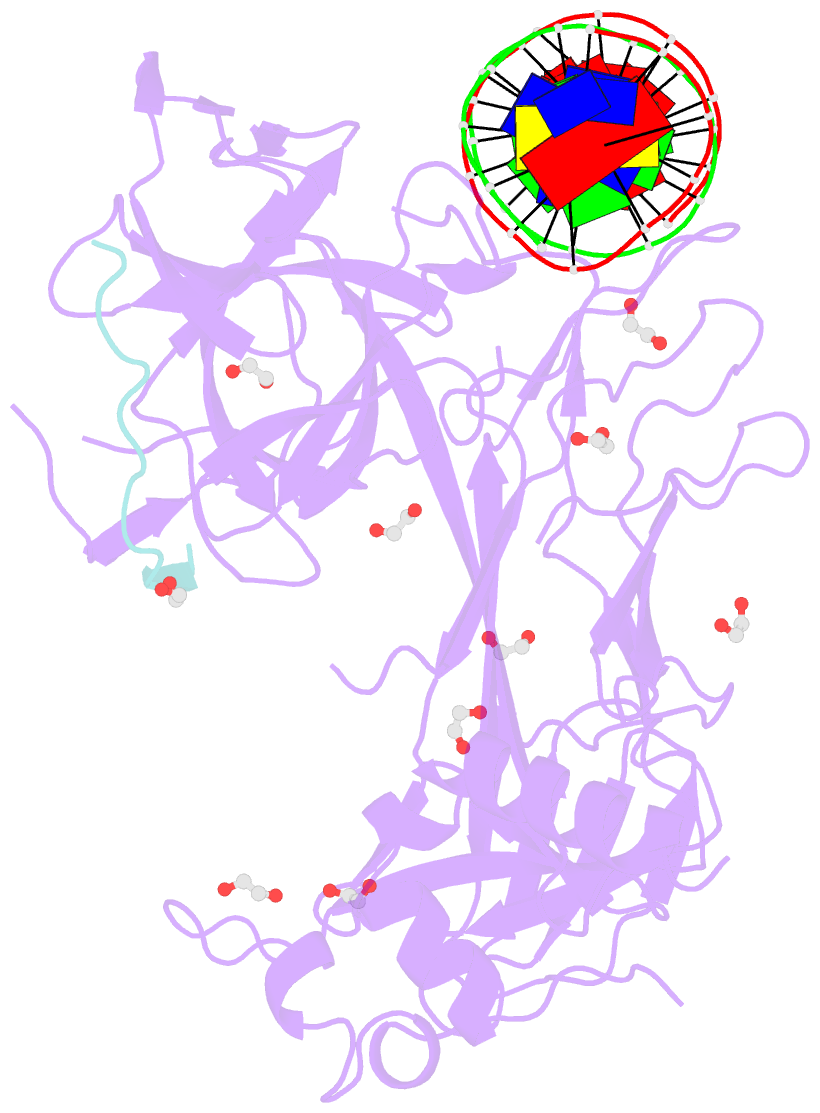
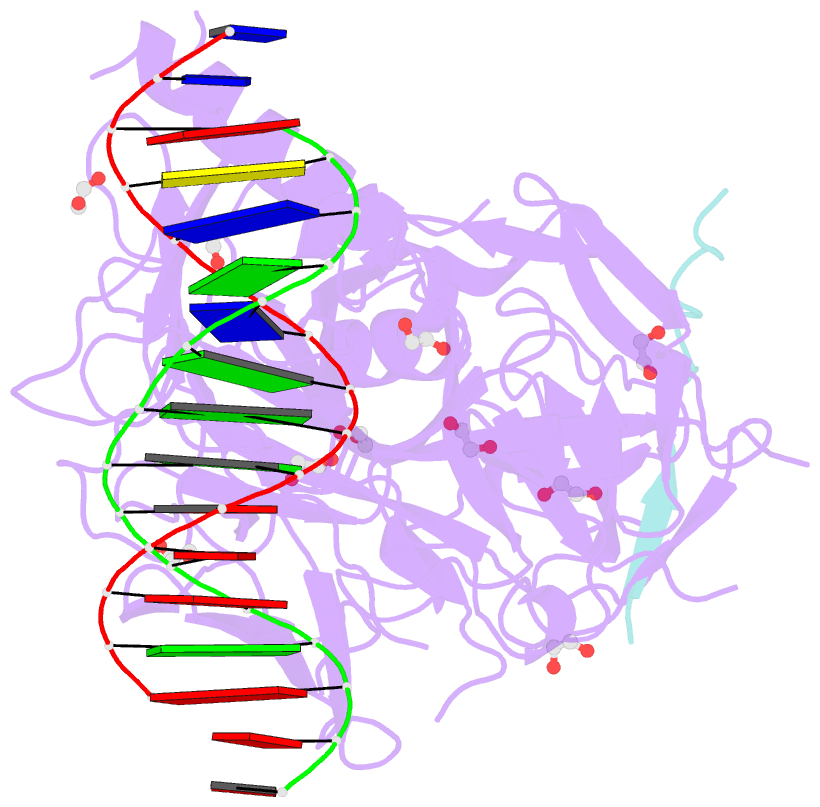
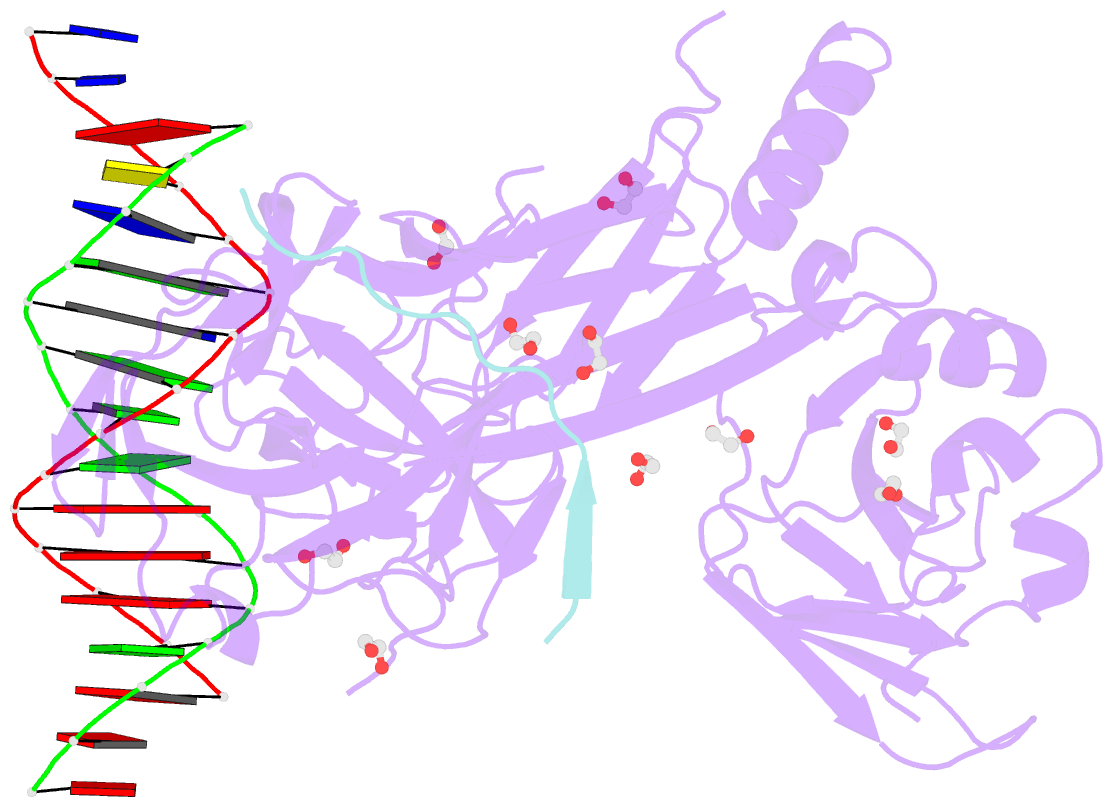
- Csl (lag-1) bound to DNA with lin-12 ram peptide, p212121
- Reference
- Friedmann DR, Wilson JJ, Kovall RA (2008): "RAM-induced Allostery Facilitates Assembly of a Notch Pathway Active Transcription Complex." J.Biol.Chem., 283, 14781-14791. doi: 10.1074/jbc.M709501200.
- Abstract
- The Notch pathway is a conserved cell-to-cell signaling mechanism, in which extracellular signals are transduced into transcriptional outputs through the nuclear effector CSL. CSL is converted from a repressor to an activator through the formation of the CSL-NotchIC-Mastermind ternary complex. The RAM (RBP-J associated molecule) domain of NotchIC avidly interacts with CSL; however, its role in assembly of the CSL-NotchIC-Mastermind ternary complex is not understood. Here we provide a comprehensive thermodynamic, structural, and biochemical analysis of the RAM-CSL interaction for components from both mouse and worm. Our binding data show that RAM and CSL form a high affinity complex in the presence or absence of DNA. Our structural studies reveal a striking distal conformational change in CSL upon RAM binding, which creates a docking site for Mastermind to bind to the complex. Finally, we show that the addition of a RAM peptide in trans facilitates formation of the CSL-NotchIC-Mastermind ternary complex in vitro.