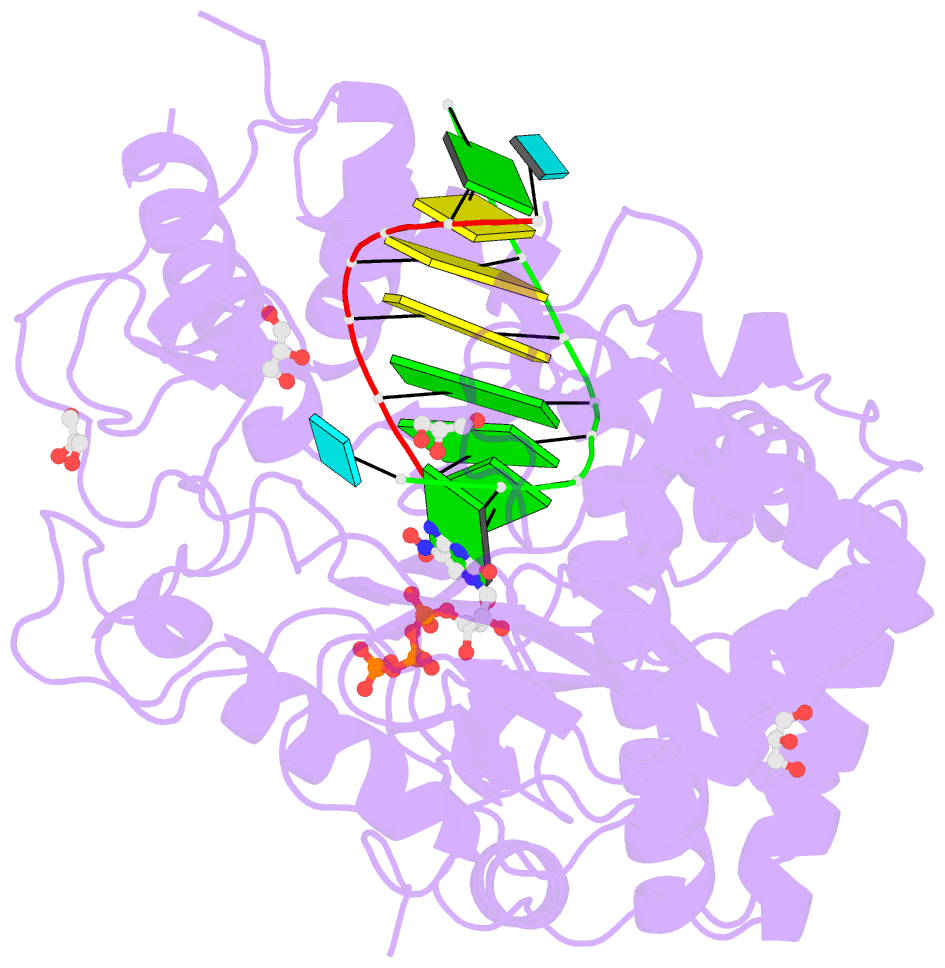
Summary information and primary citation
- PDB-id
- 3bsn; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-RNA
- Method
- X-ray (1.8 Å)
- Summary
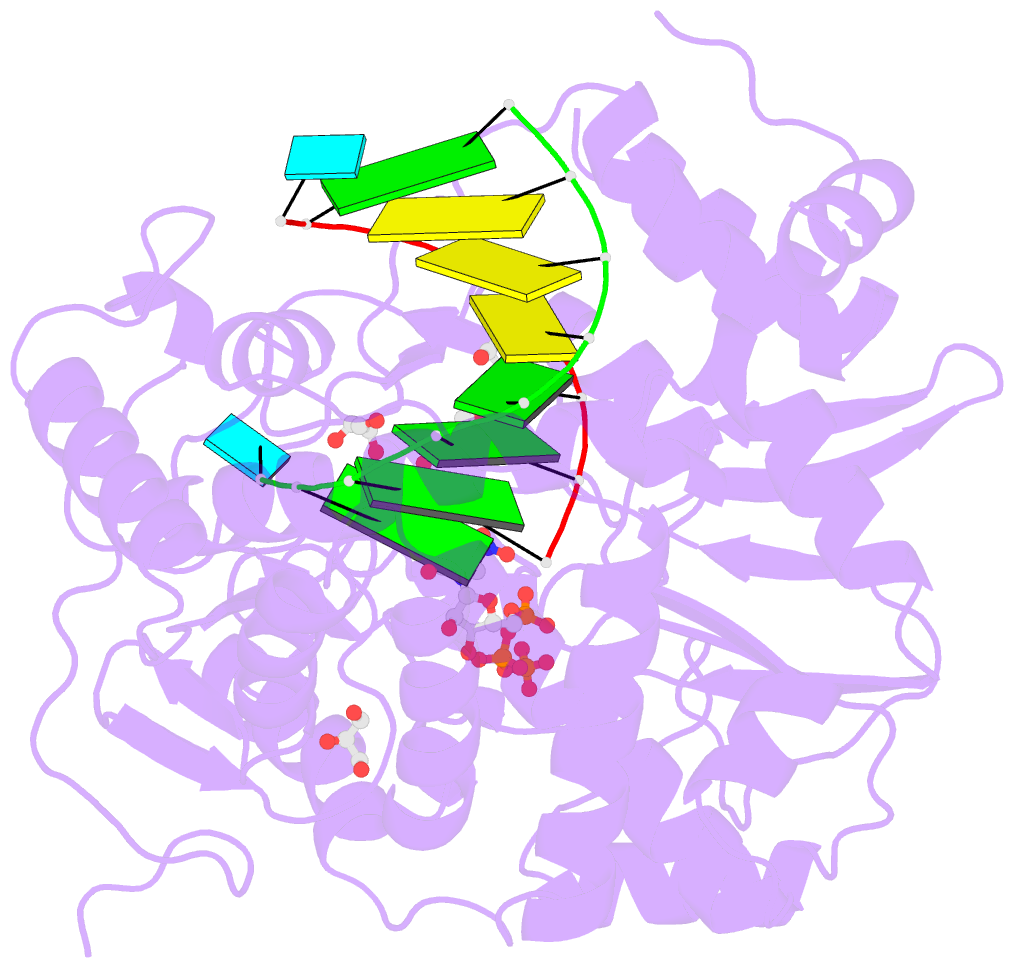
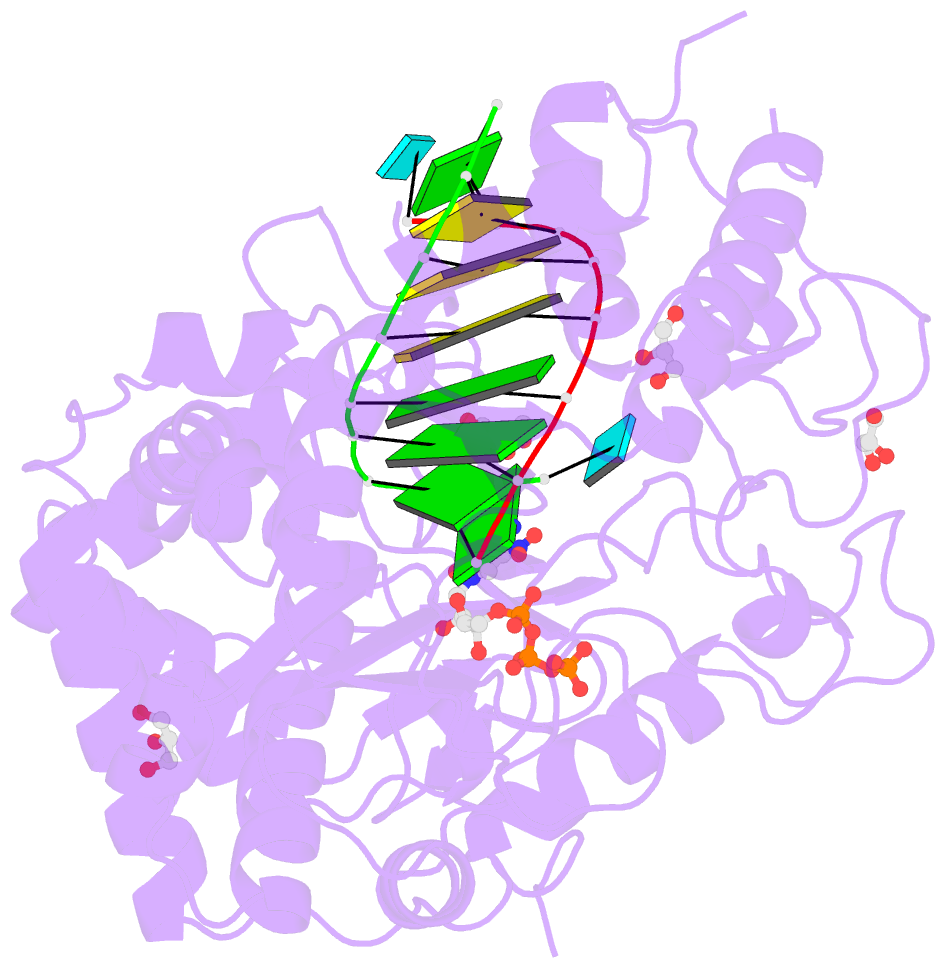
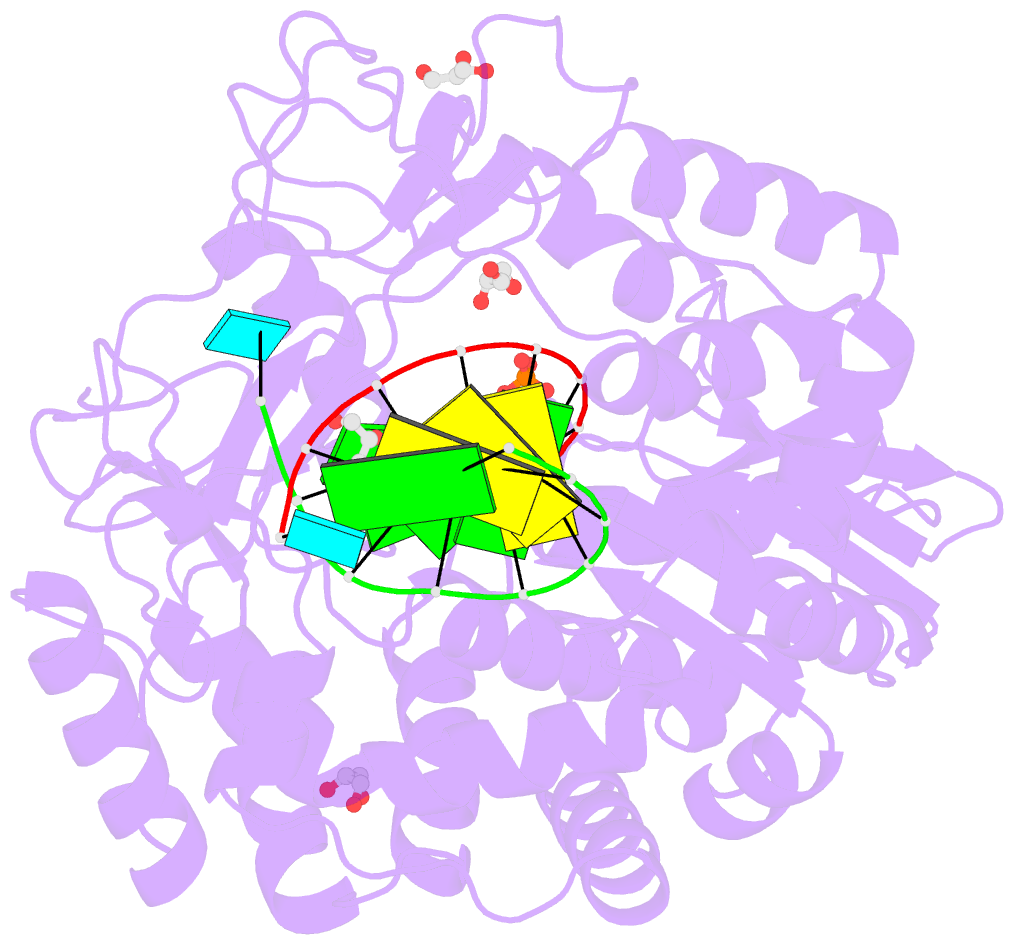
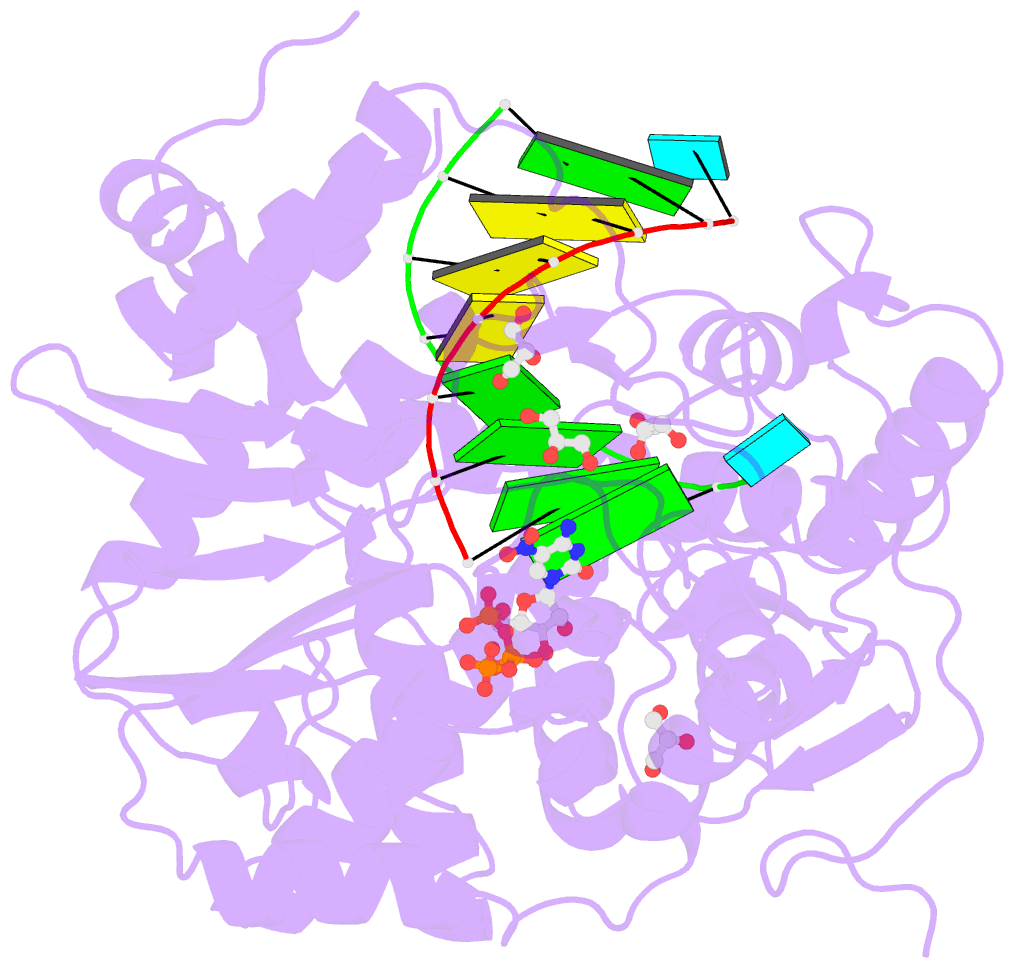
- Norwalk virus polymerase bound to 5-nitrocytidine triphosphate and primer-template RNA
- Reference
- Zamyatkin DF, Parra F, Alonso JM, Harki DA, Peterson BR, Grochulski P, Ng KK (2008): "Structural insights into mechanisms of catalysis and inhibition in norwalk virus polymerase." J.Biol.Chem., 283, 7705-7712. doi: 10.1074/jbc.M709563200.
- Abstract
- Crystal structures of Norwalk virus polymerase bound to an RNA primer-template duplex and either the natural substrate CTP or the inhibitor 5-nitrocytidine triphosphate have been determined to 1.8A resolution. These structures reveal a closed conformation of the polymerase that differs significantly from previously determined open structures of calicivirus and picornavirus polymerases. These closed complexes are trapped immediately prior to the nucleotidyl transfer reaction, with the triphosphate group of the nucleotide bound to two manganese ions at the active site, poised for reaction to the 3'-hydroxyl group of the RNA primer. The positioning of the 5-nitrocytidine triphosphate nitro group between the alpha-phosphate and the 3'-hydroxyl group of the primer suggests a novel, general approach for the design of antiviral compounds mimicking natural nucleosides and nucleotides.