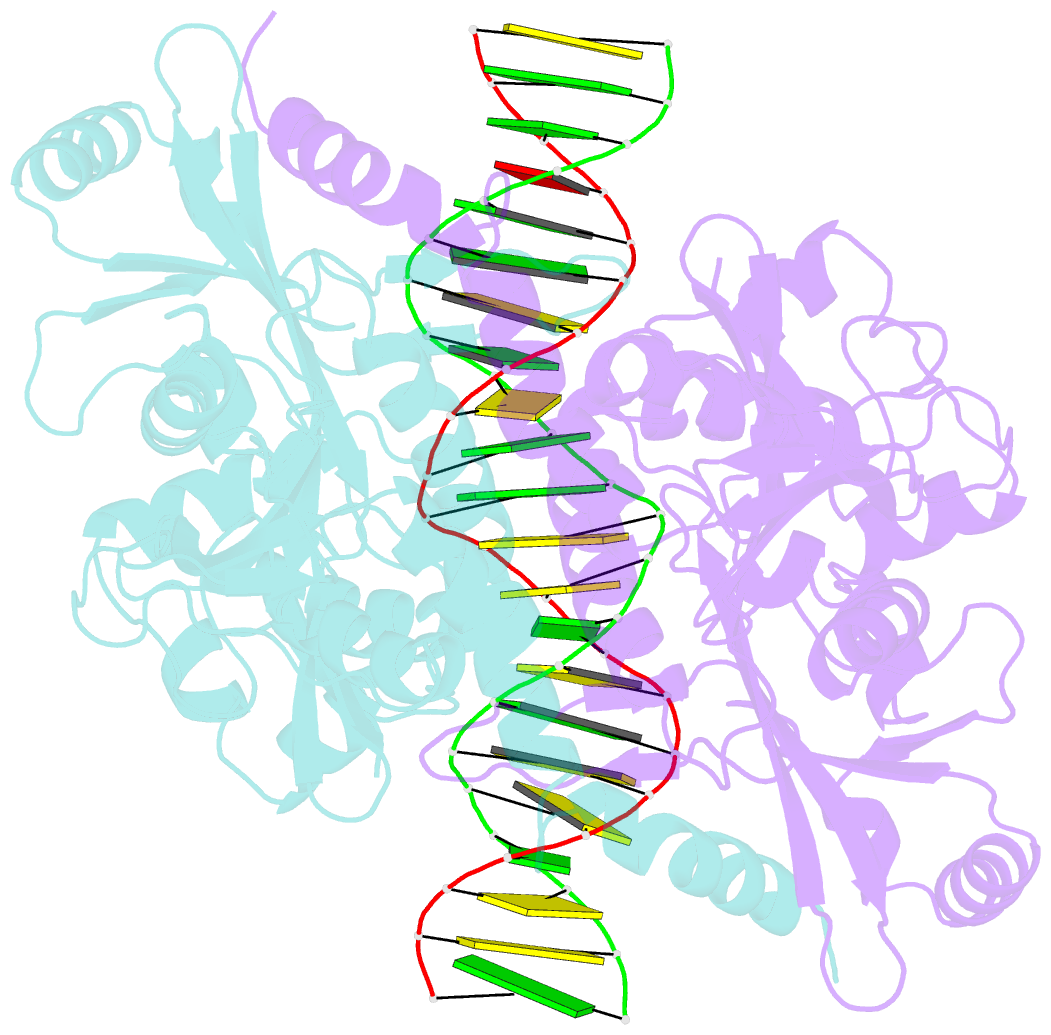
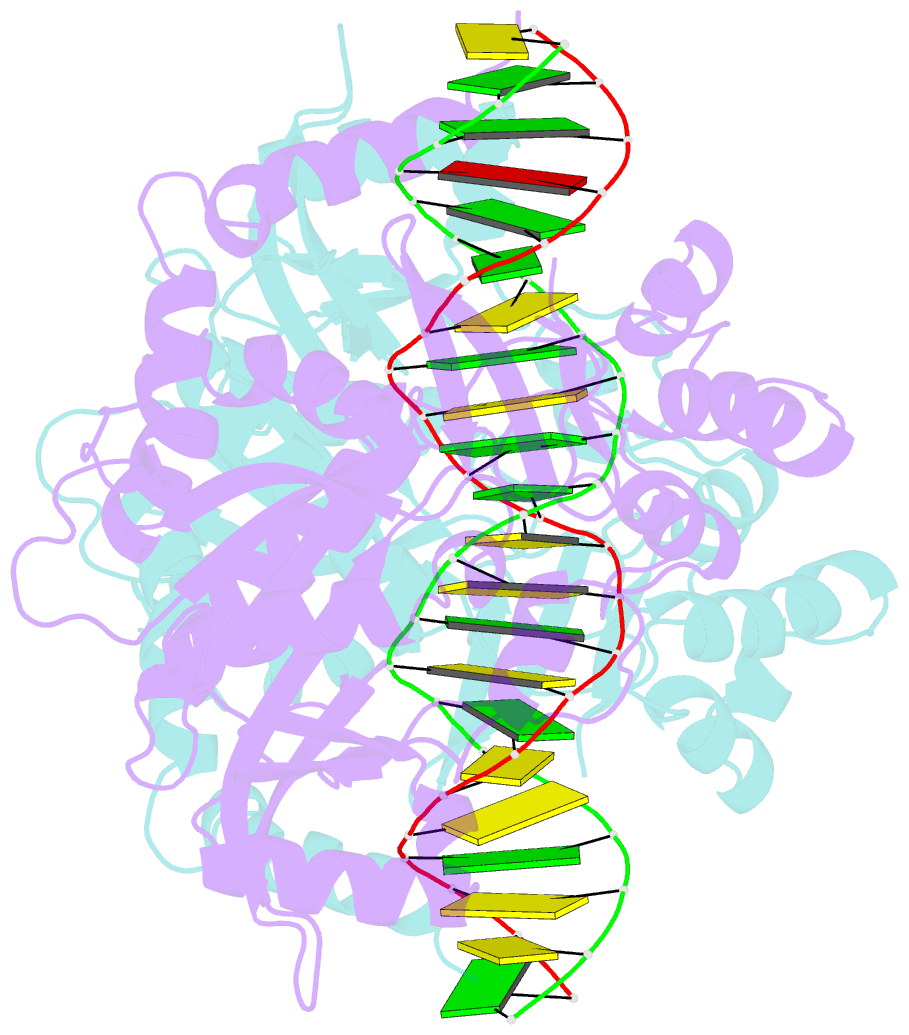
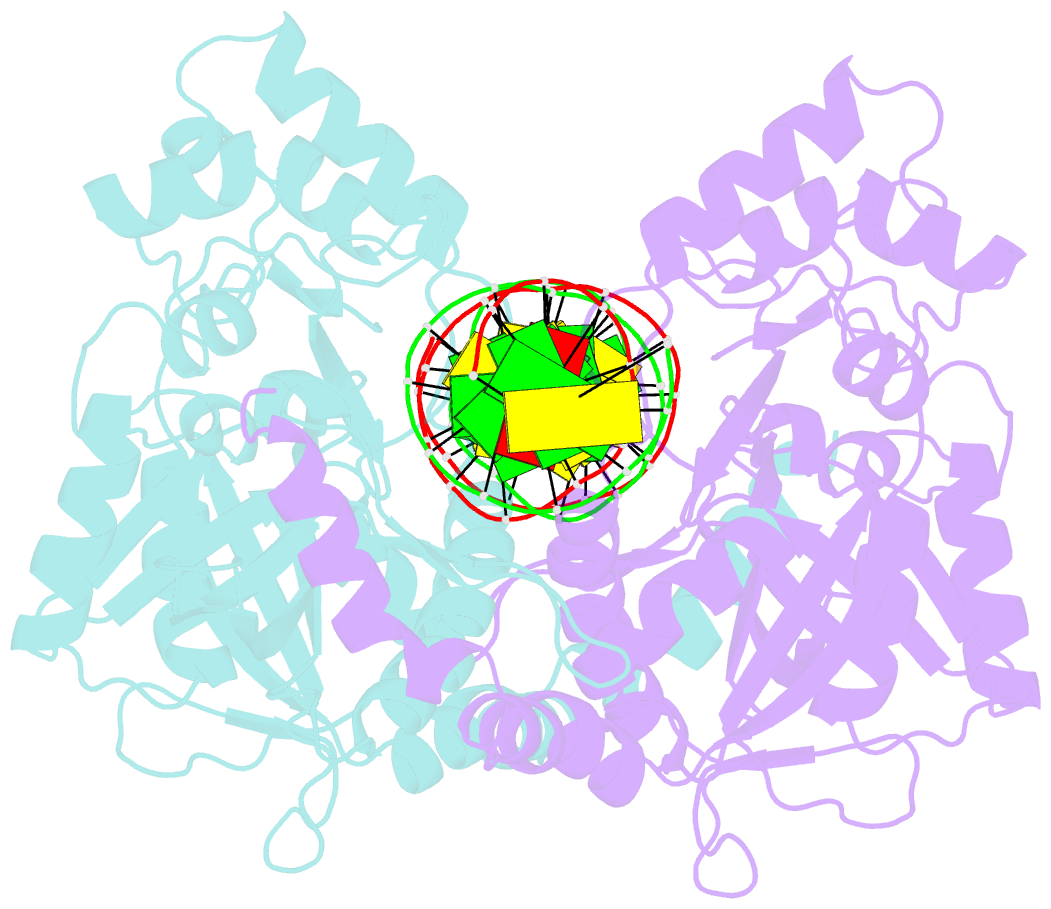
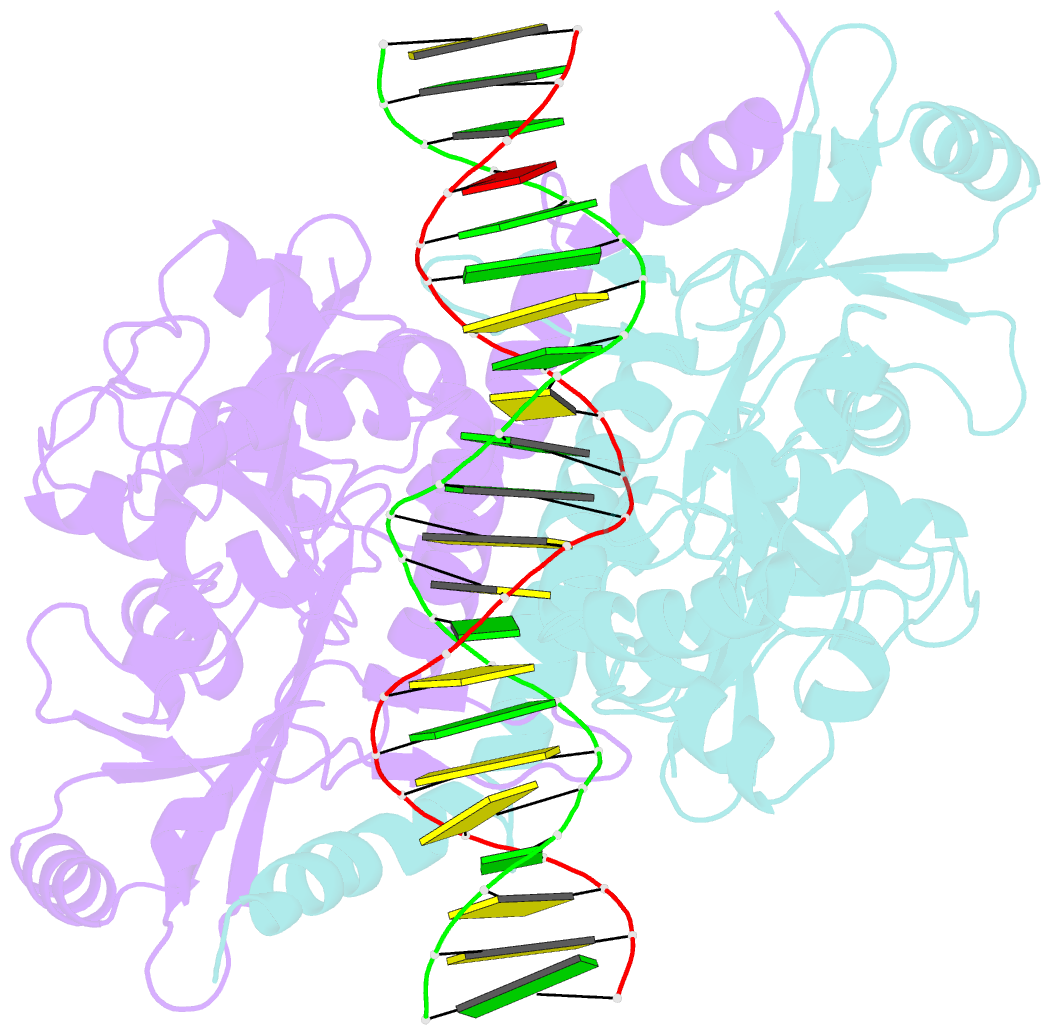
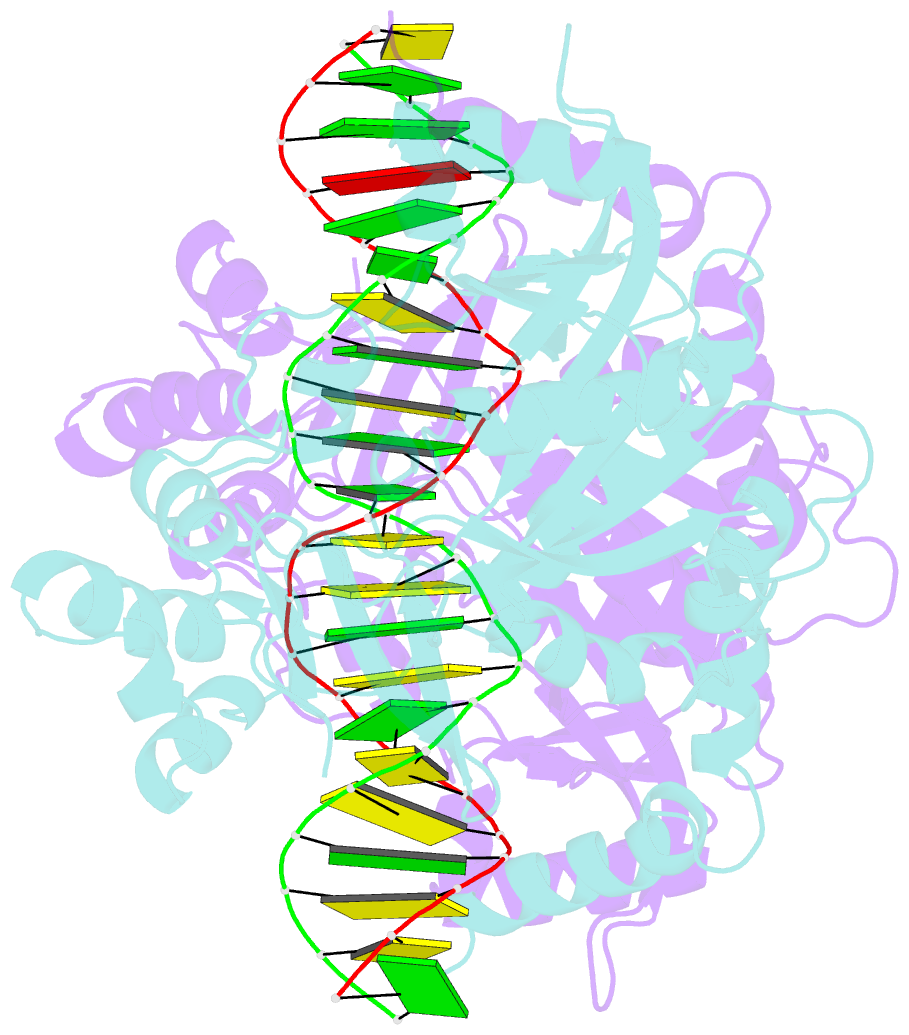
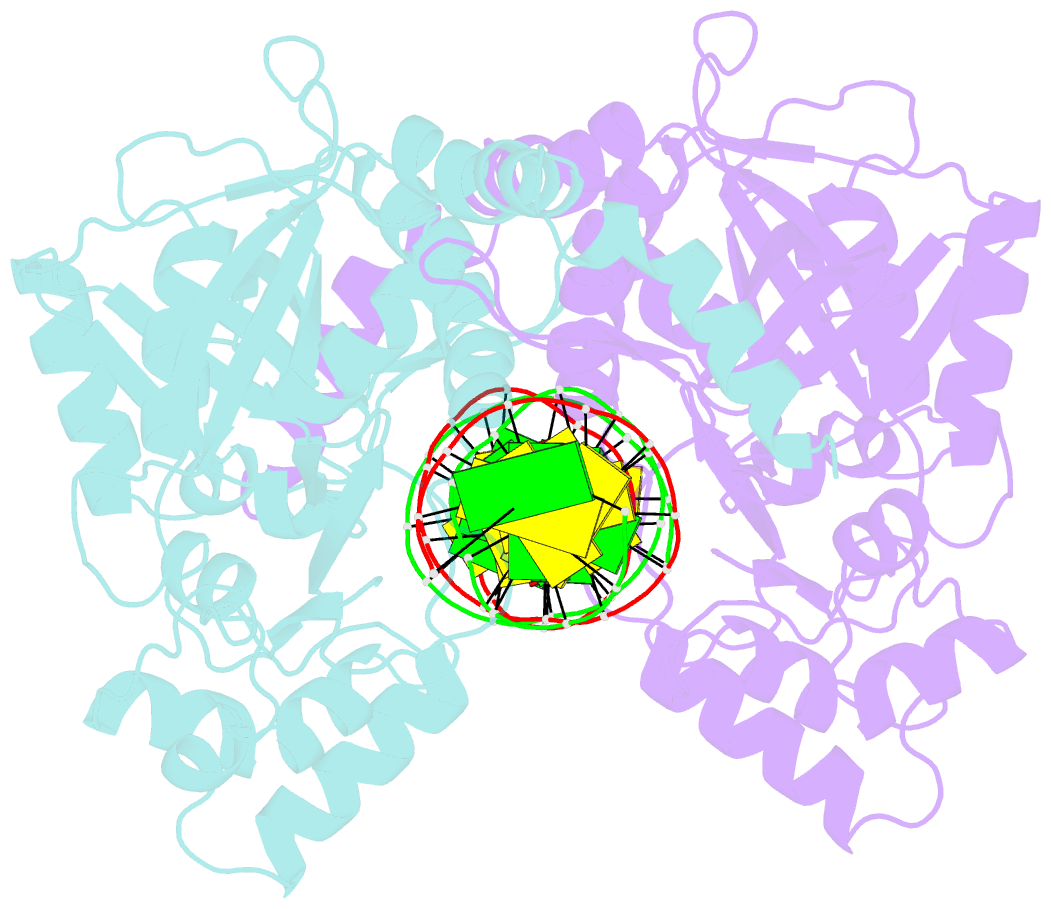
Summary information and primary citation
- PDB-id
- 3c25; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.5 Å)
- Summary
- Crystal structure of noti restriction endonuclease bound to cognate DNA
- Reference
- Lambert AR, Sussman D, Shen B, Maunus R, Nix J, Samuelson J, Xu SY, Stoddard BL (2008): "Structures of the Rare-Cutting Restriction Endonuclease NotI Reveal a Unique Metal Binding Fold Involved in DNA Binding." Structure, 16, 558-569. doi: 10.1016/j.str.2008.01.017.
- Abstract
- The structure of the rare-cutting restriction endonuclease NotI, which recognizes the 8 bp target 5'-GCGGCCGC-3', has been solved with and without bound DNA. Because of its specificity (recognizing a site that occurs once per 65 kb), NotI is used to generate large genomic fragments and to map DNA methylation status. NotI contains a unique metal binding fold, found in a variety of putative endonucleases, occupied by an iron atom coordinated within a tetrahedral Cys4 motif. This domain positions nearby protein elements for DNA recognition, and serves a structural role. While recognition of the central six base pairs of the target is accomplished via a saturated hydrogen bond network typical of restriction enzymes, the most peripheral base pairs are engaged in a single direct contact in the major groove, reflecting reduced pressure to recognize those positions. NotI may represent an evolutionary intermediate between mobile endonucleases (which recognize longer target sites) and canonical restriction endonucleases.