Summary information and primary citation
- PDB-id
- 3d2w; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA-RNA binding protein
- Method
- X-ray (1.65 Å)
- Summary
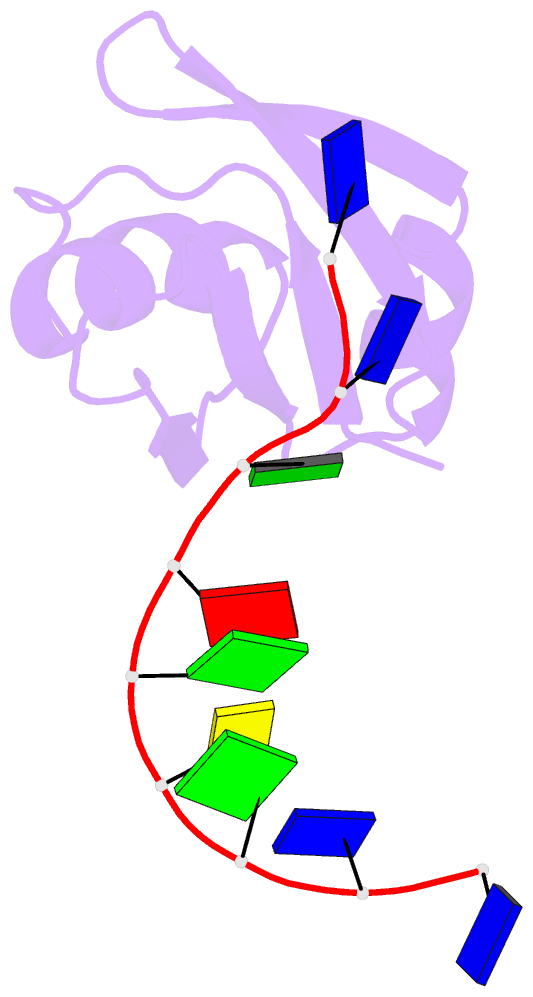
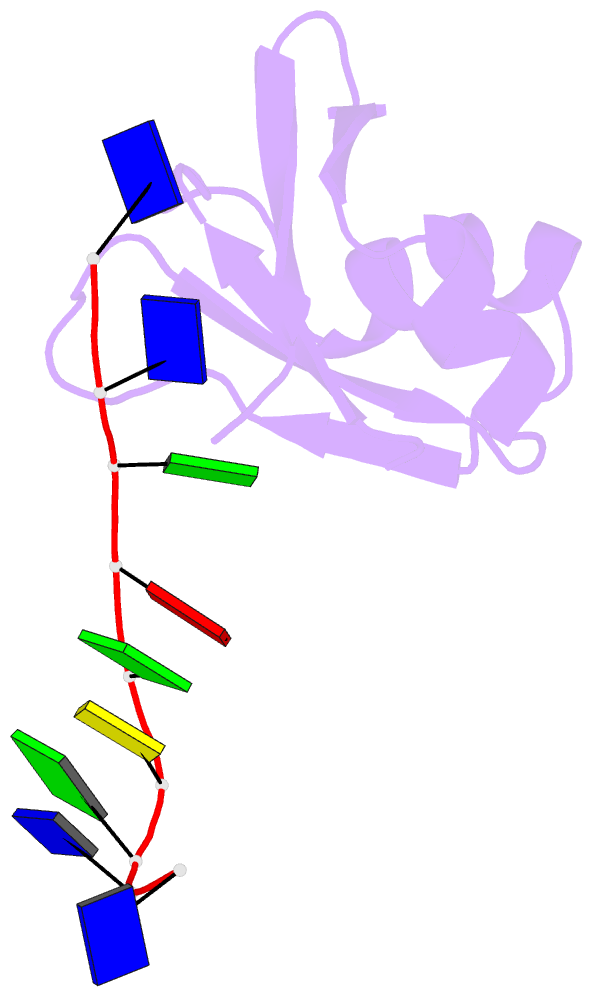
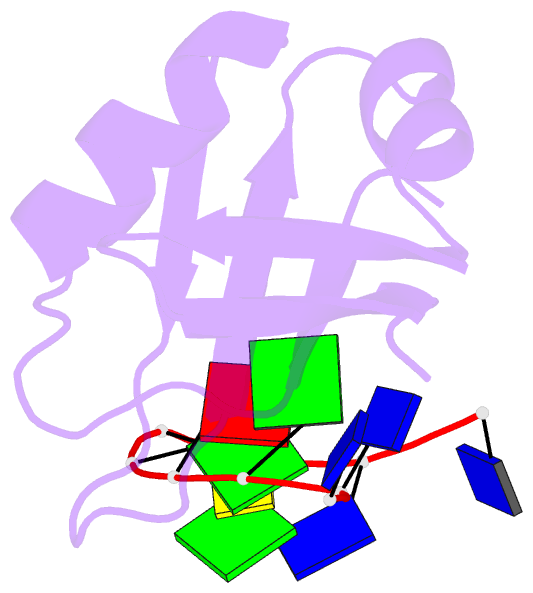
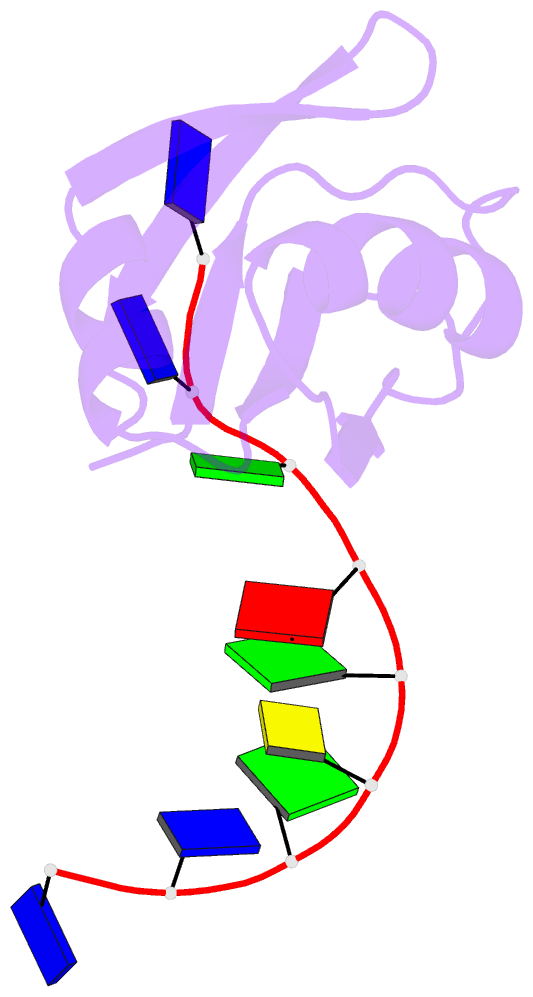
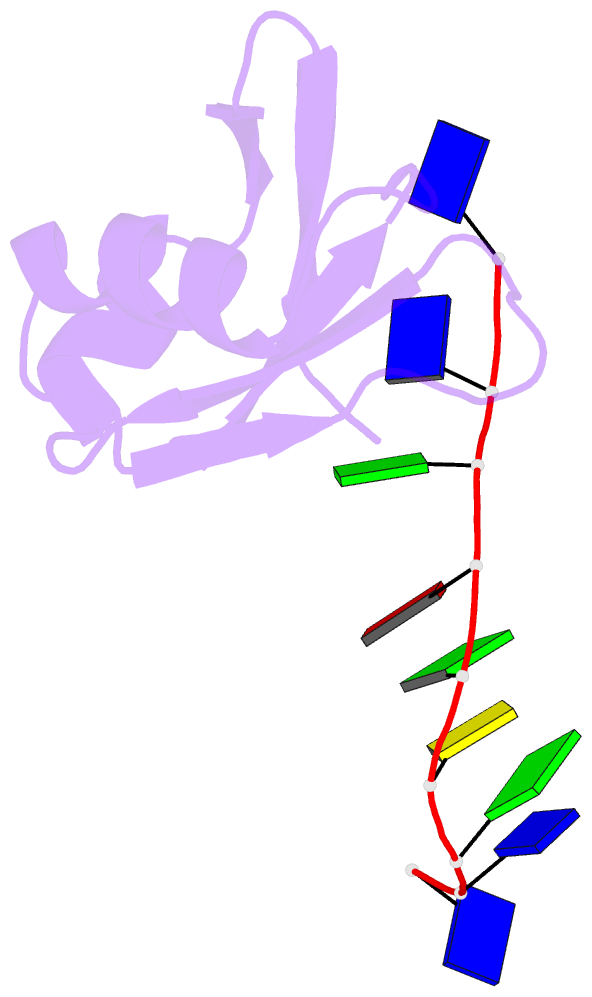
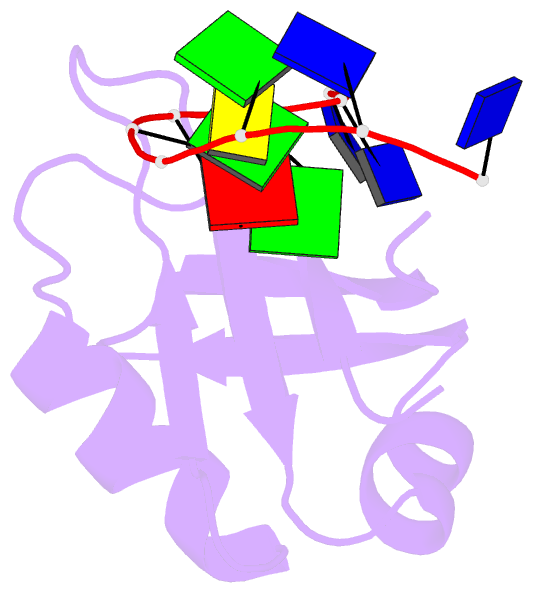
- Crystal structure of mouse tdp-43 rrm2 domain in complex with DNA
- Reference
- Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS (2009): "Structural insights into TDP-43 in nucleic-acid binding and domain interactions." Nucleic Acids Res., 37, 1799-1808. doi: 10.1093/nar/gkp013.
- Abstract
- TDP-43 is a pathogenic protein: its normal function in binding to UG-rich RNA is related to cystic fibrosis, and inclusion of its C-terminal fragments in brain cells is directly linked to frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS). Here we report the 1.65 A crystal structure of the C-terminal RRM2 domain of TDP-43 in complex with a single-stranded DNA. We show that TDP-43 is a dimeric protein with two RRM domains, both involved in DNA and RNA binding. The crystal structure reveals the basis of TDP-43's TG/UG preference in nucleic acids binding. It also reveals that RRM2 domain has an atypical RRM-fold with an additional beta-strand involved in making protein-protein interactions. This self association of RRM2 domains produced thermal-stable RRM2 assemblies with a melting point greater than 85 degrees C as monitored by circular dichroism at physiological conditions. These studies thus characterize the recognition between TDP-43 and nucleic acids and the mode of RRM2 self association, and provide molecular models for understanding the role of TDP-43 in cystic fibrosis and the neurodegenerative diseases related to TDP-43 proteinopathy.