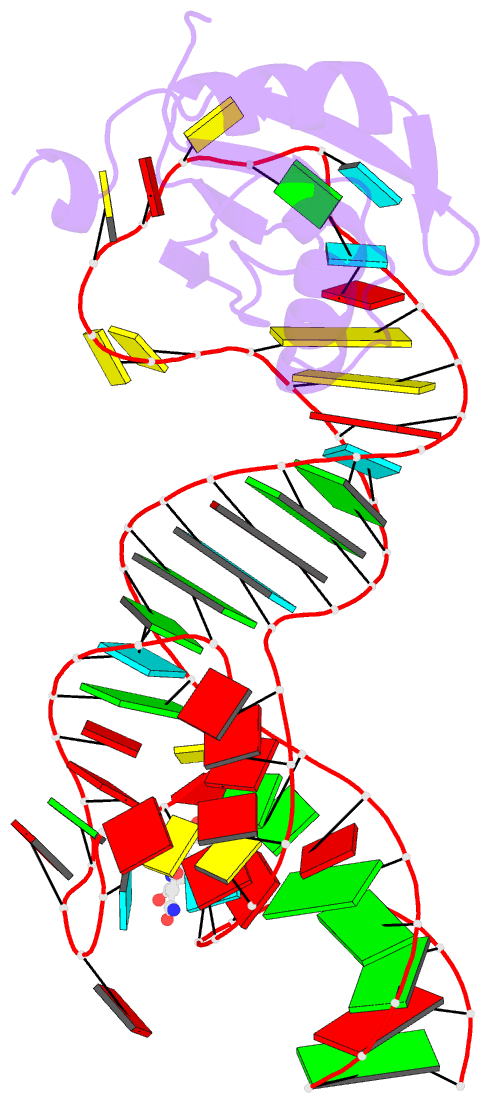
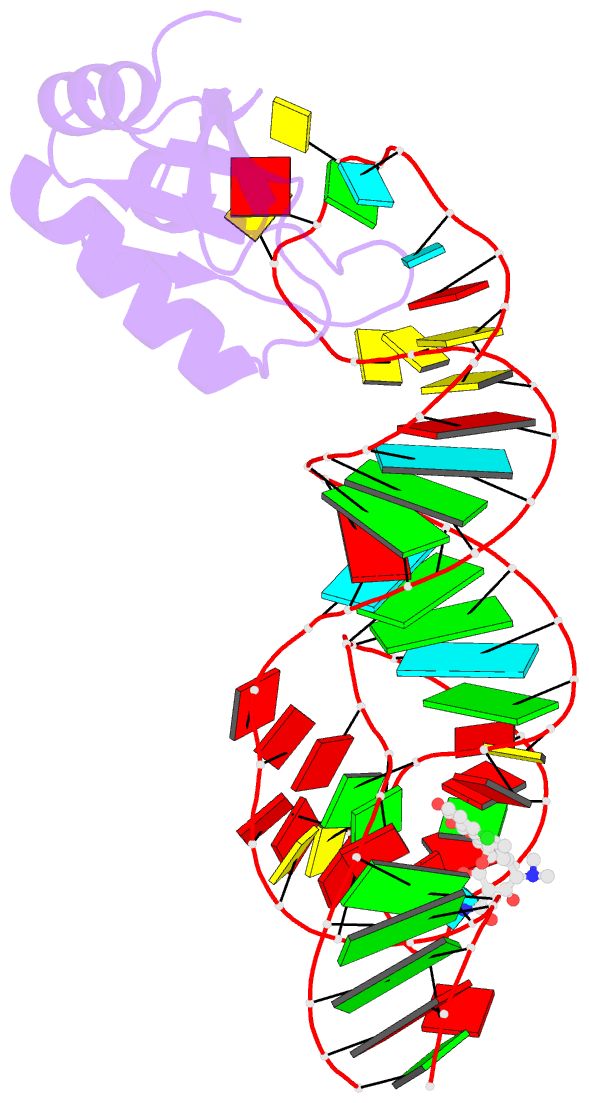
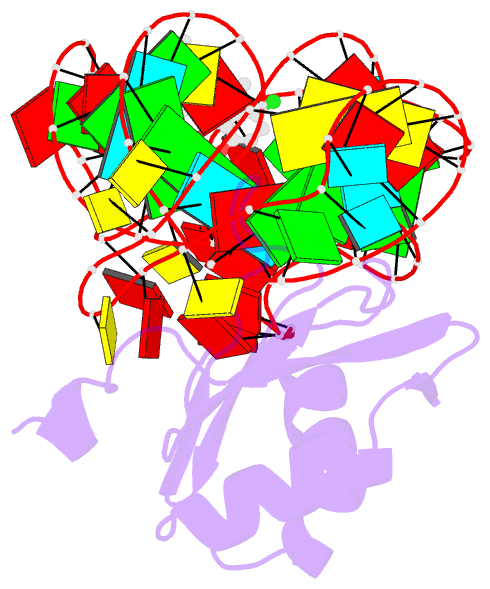
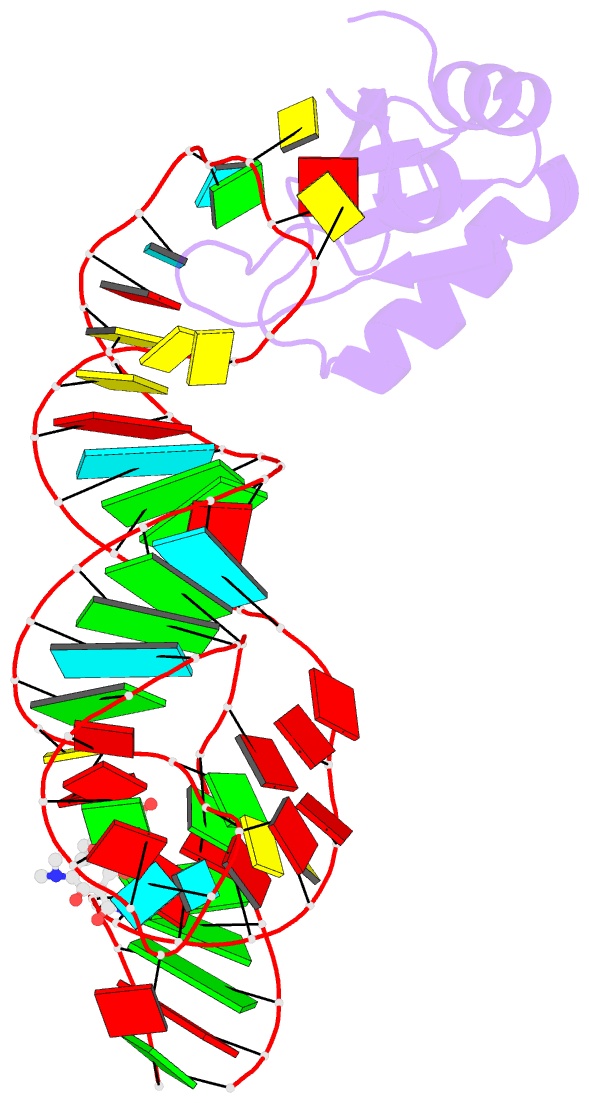
Summary information and primary citation
- PDB-id
- 3egz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA
- Method
- X-ray (2.2 Å)
- Summary
- Crystal structure of an in vitro evolved tetracycline aptamer and artificial riboswitch
- Reference
- Xiao H, Edwards TE, Ferre-D'Amare AR (2008): "Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch." Chem.Biol., 15, 1125-1137. doi: 10.1016/j.chembiol.2008.09.004.
- Abstract
- The tetracycline aptamer is an in vitro selected RNA that binds to the antibiotic with the highest known affinity of an artificial RNA for a small molecule (Kd approximately 0.8 nM). It is one of few aptamers known to be capable of modulating gene expression in vivo. The 2.2 A resolution cocrystal structure of the aptamer reveals a pseudoknot-like fold formed by tertiary interactions between an 11 nucleotide loop and the minor groove of an irregular helix. Tetracycline binds within this interface as a magnesium ion chelate. The structure, together with previous biochemical and biophysical data, indicates that the aptamer undergoes localized folding concomitant with tetracycline binding. The three-helix junction, h-shaped architecture of this artificial RNA is more complex than those of most aptamers and is reminiscent of the structures of some natural riboswitches.