Summary information and primary citation
- PDB-id
- 3er9; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription, transferase-RNA
- Method
- X-ray (2.06 Å)
- Summary
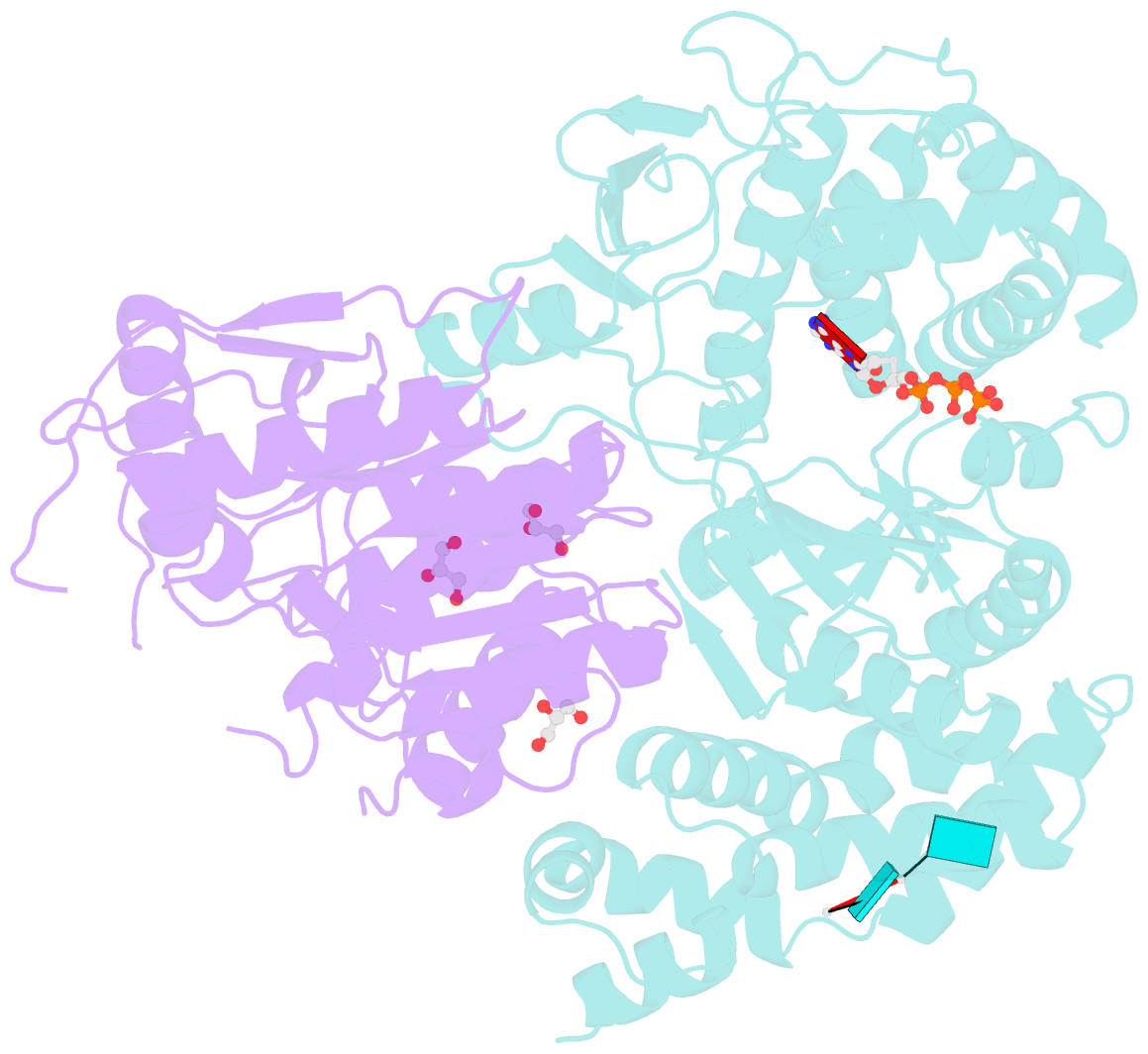
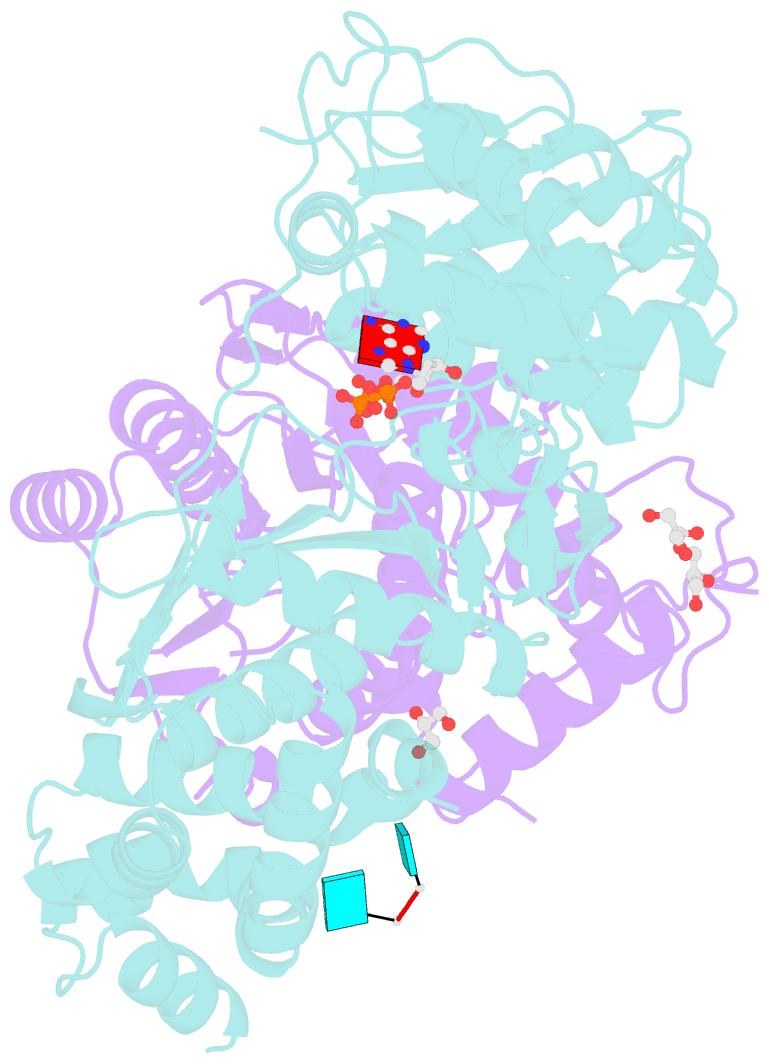
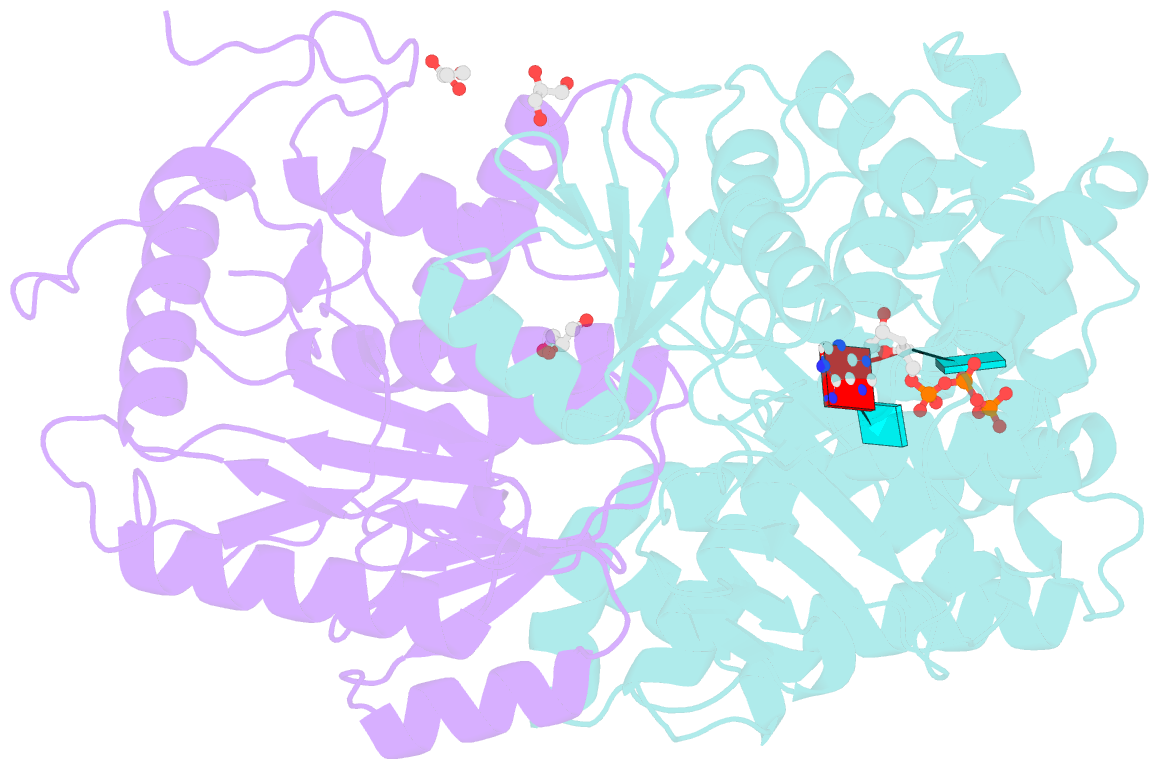
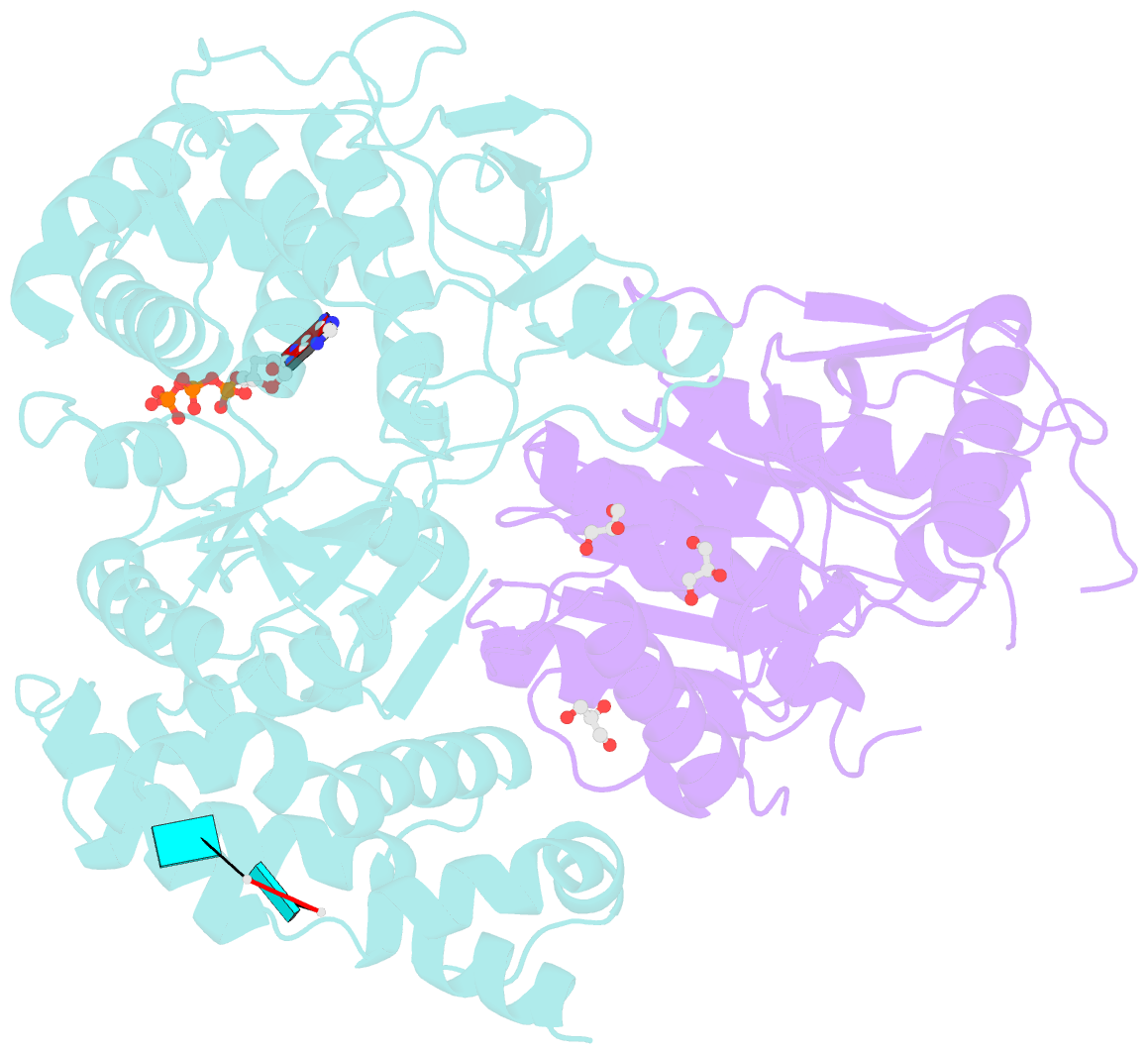
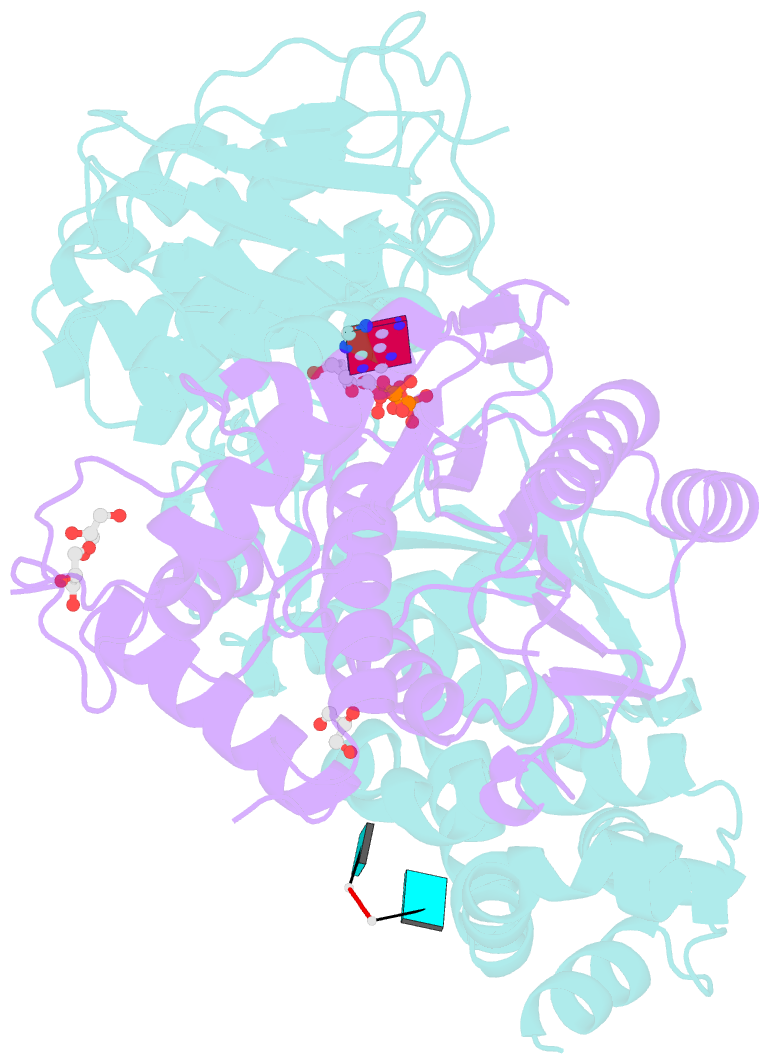
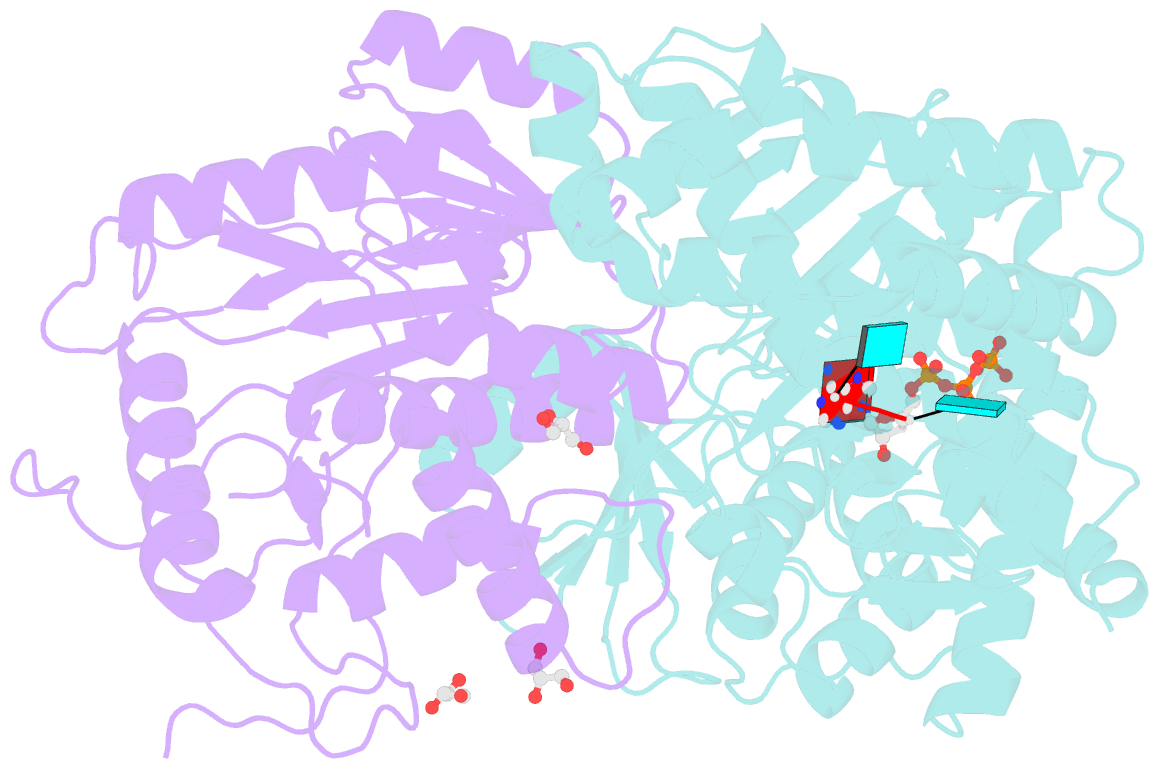
- Crystal structure of the heterodimeric vaccinia virus mrna polyadenylate polymerase complex with uu and 3'-deoxy atp
- Reference
- Li C, Li H, Zhou S, Sun E, Yoshizawa J, Poulos TL, Gershon PD (2009): "Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase." Structure, 17, 680-689. doi: 10.1016/j.str.2009.03.012.
- Abstract
- Vaccinia virus protein VP55 translocates continuously with respect to single-stranded nucleic acid while extending its 3'end. Here, all key sites of polymerase-primer interaction were identified, demonstrating the wrapping or looping of polyadenylation primer around the polymerase during translocation. Side-chain substitutions at one of the sites indicated its requirement for tail extension beyond approximately 12 nucleotides in length, and conformational changes observed upon oligonucleotide binding suggested allosteric connectivity during translocation. Conformational changes in VP39 upon VP55 binding suggested that, within the VP55-VP39 complex, VP39's mRNA 5' cap binding site closes. The crystallographic structure showed a PAPase catalytic center without side-chain substitutions, possessing two metal ions and with all known reactive and catalytic groups represented, fitting a classical two-metal ion mechanism for phosphoryl transfer.