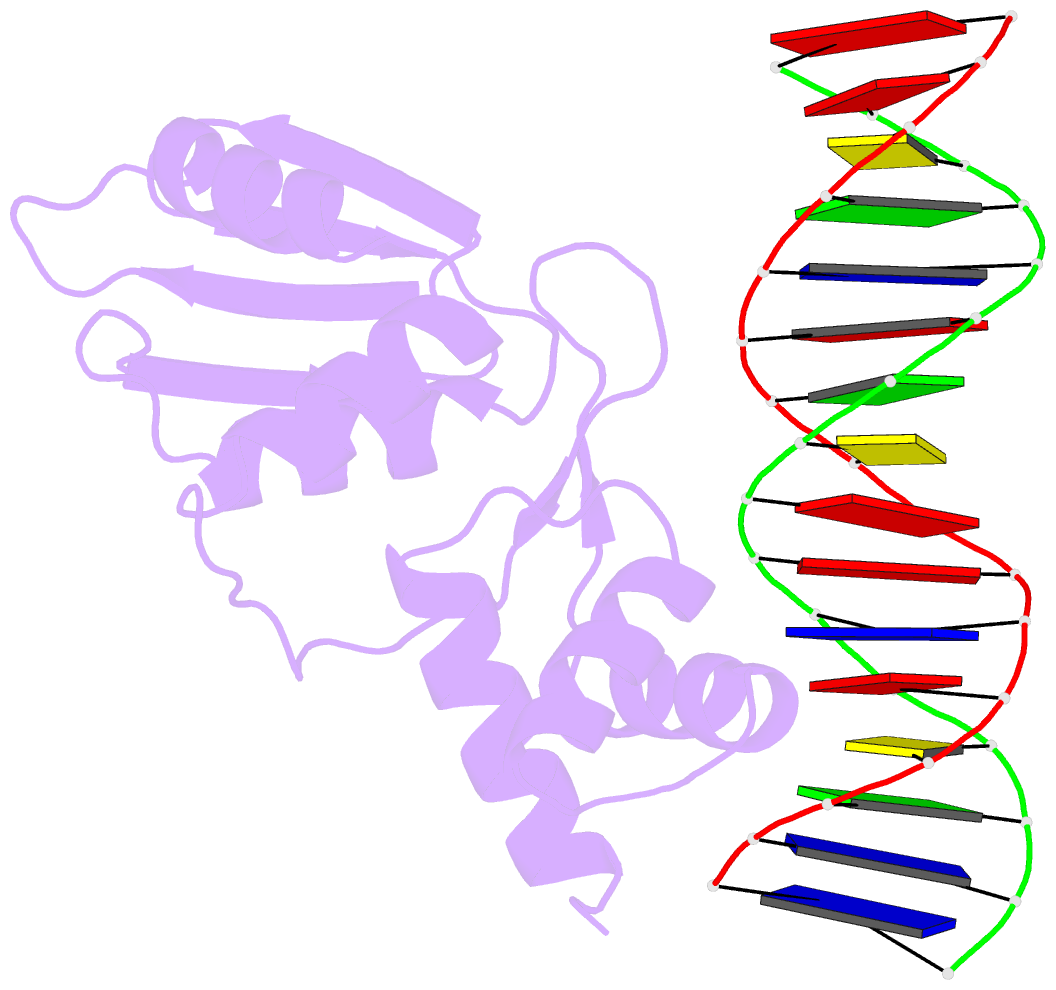
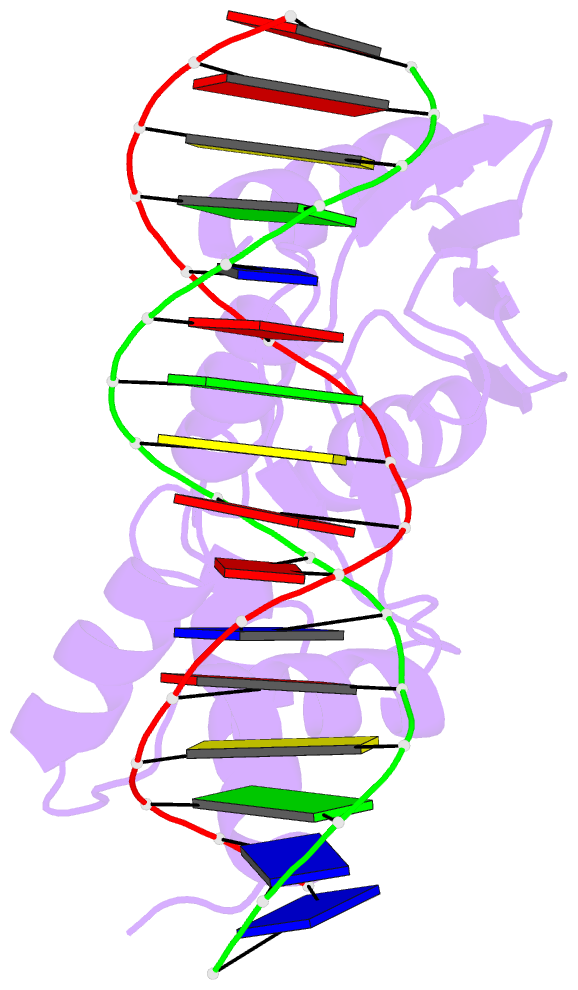
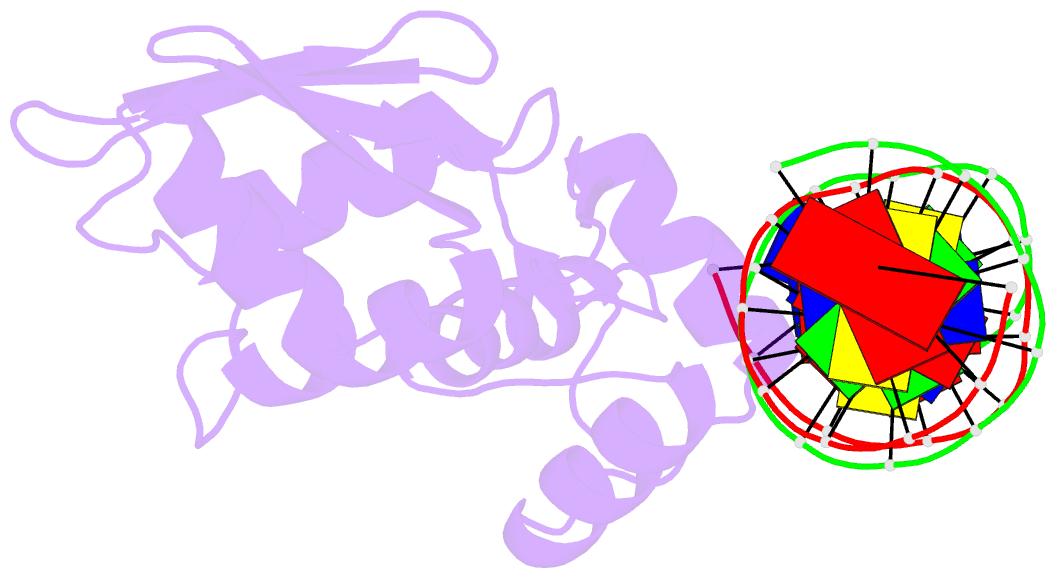
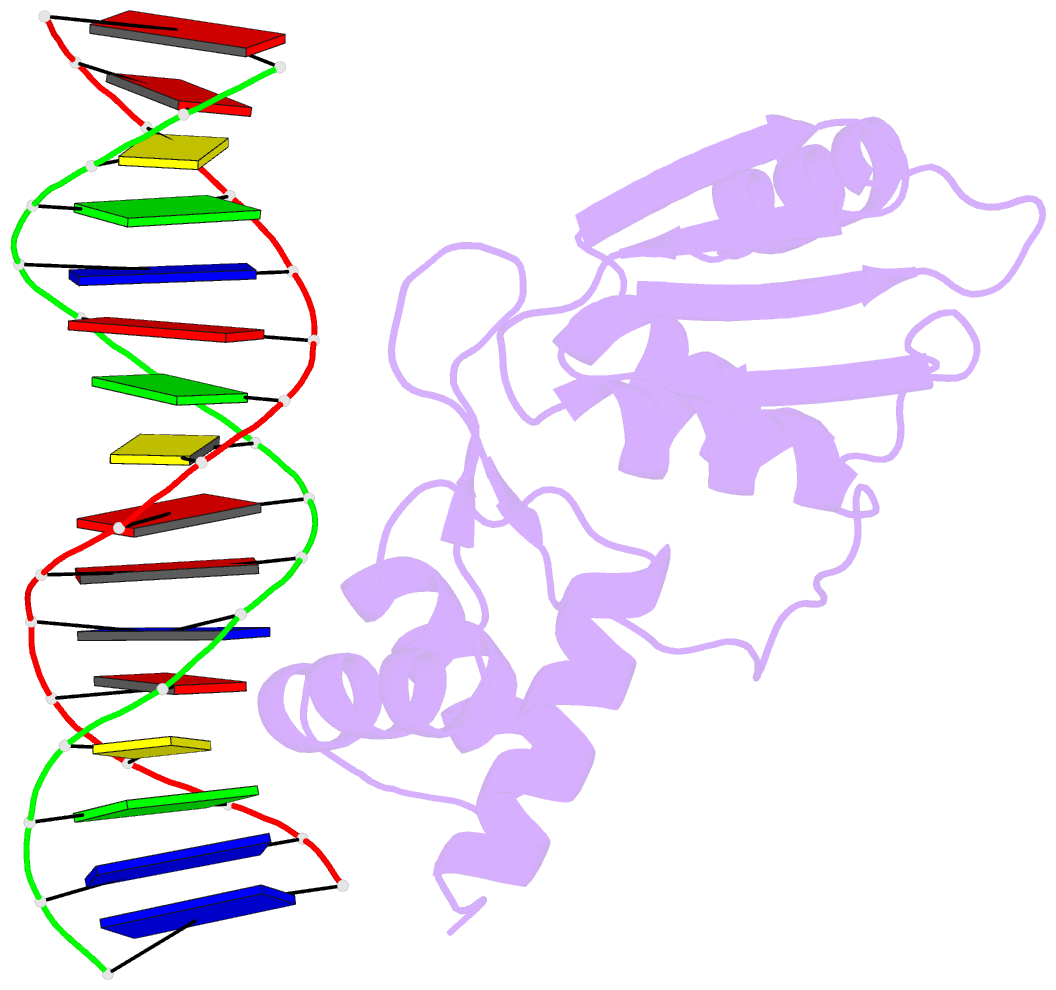
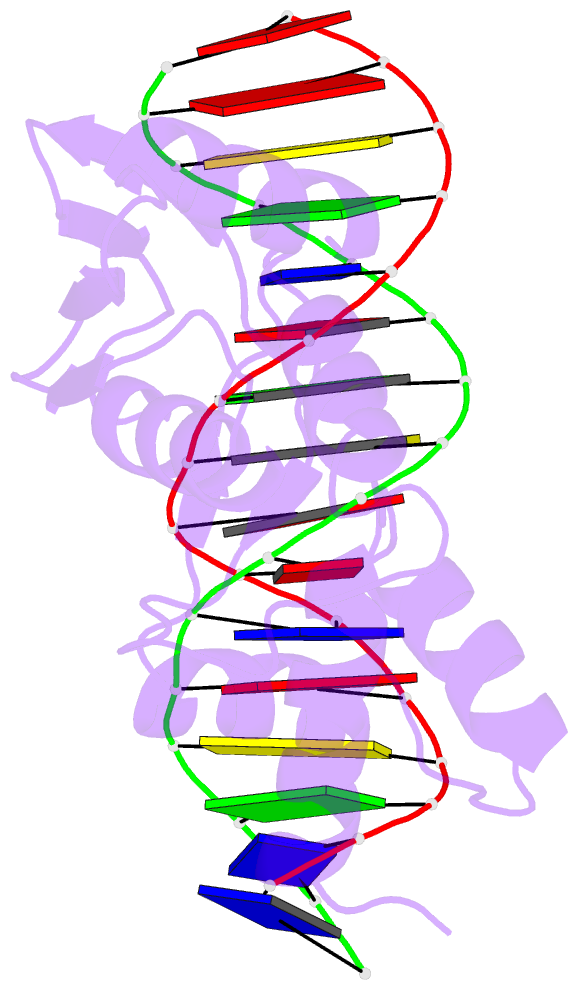
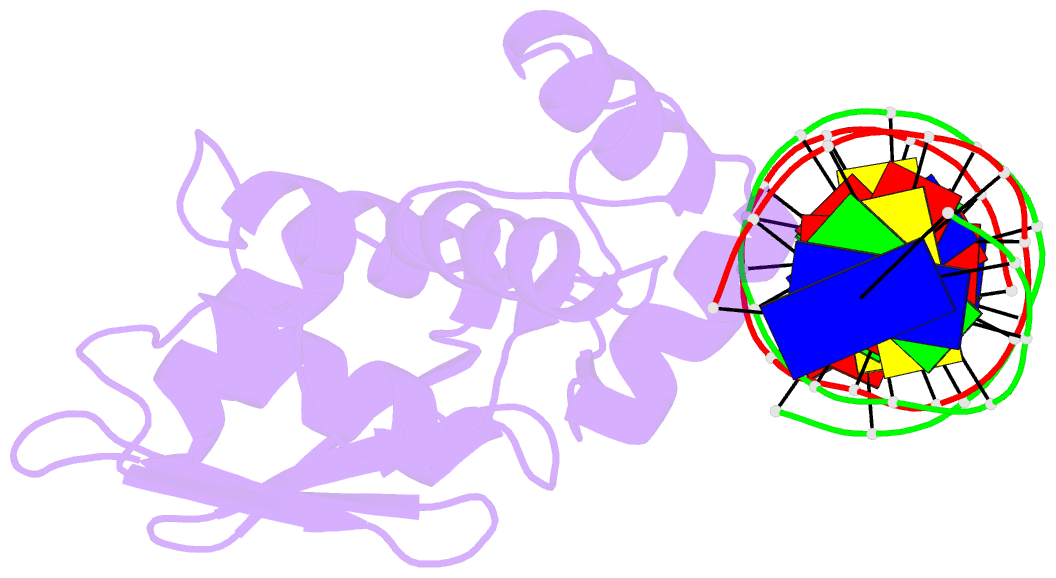
Summary information and primary citation
- PDB-id
- 3ere; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.5 Å)
- Summary
- Crystal structure of the arginine repressor protein from mycobacterium tuberculosis in complex with the DNA operator
- Reference
- Cherney LT, Cherney MM, Garen CR, Lu GJ, James MN (2008): "Crystal structure of the arginine repressor protein in complex with the DNA operator from Mycobacterium tuberculosis." J.Mol.Biol., 384, 1330-1340. doi: 10.1016/j.jmb.2008.10.015.
- Abstract
- The arginine repressor (ArgR) from Mycobacterium tuberculosis (Mtb) is a gene product encoded by the open reading frame Rv1657. It regulates the L-arginine concentration in cells by interacting with ARG boxes in the promoter regions of the arginine biosynthesis and catabolism operons. Here we present a 2.5-A structure of MtbArgR in complex with a 16-bp DNA operator in the absence of arginine. A biological trimer of the protein-DNA complex is formed via the crystallographic 3-fold symmetry axis. The N-terminal domain of MtbArgR has a winged helix-turn-helix motif that binds to the major groove of the DNA. This structure shows that, in the absence of arginine, the ArgR trimer can bind three ARG box half-sites. It also reveals the structure of the whole MtbArgR molecule itself containing both N-terminal and C-terminal domains.