Summary information and primary citation
- PDB-id
- 3fsi; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (1.75 Å)
- Summary
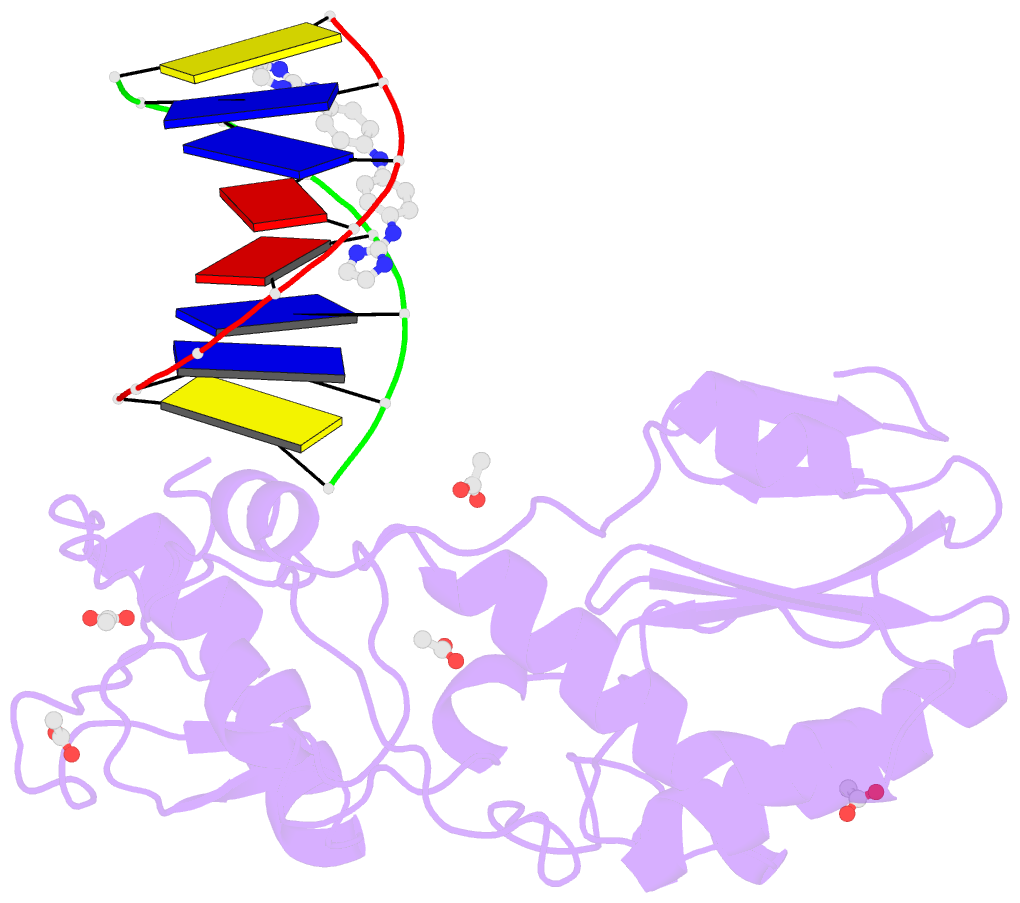
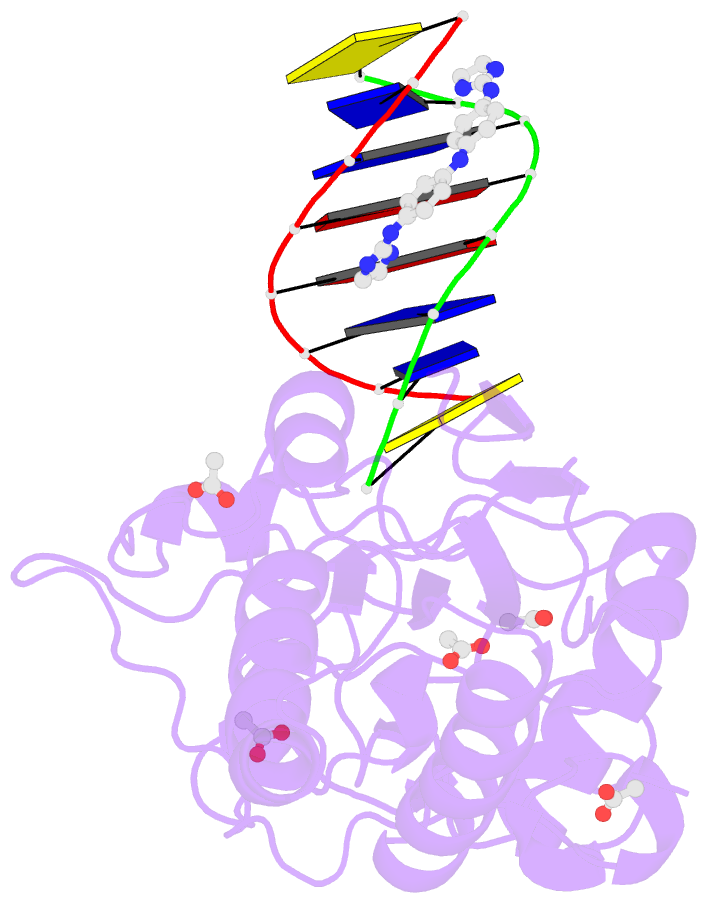
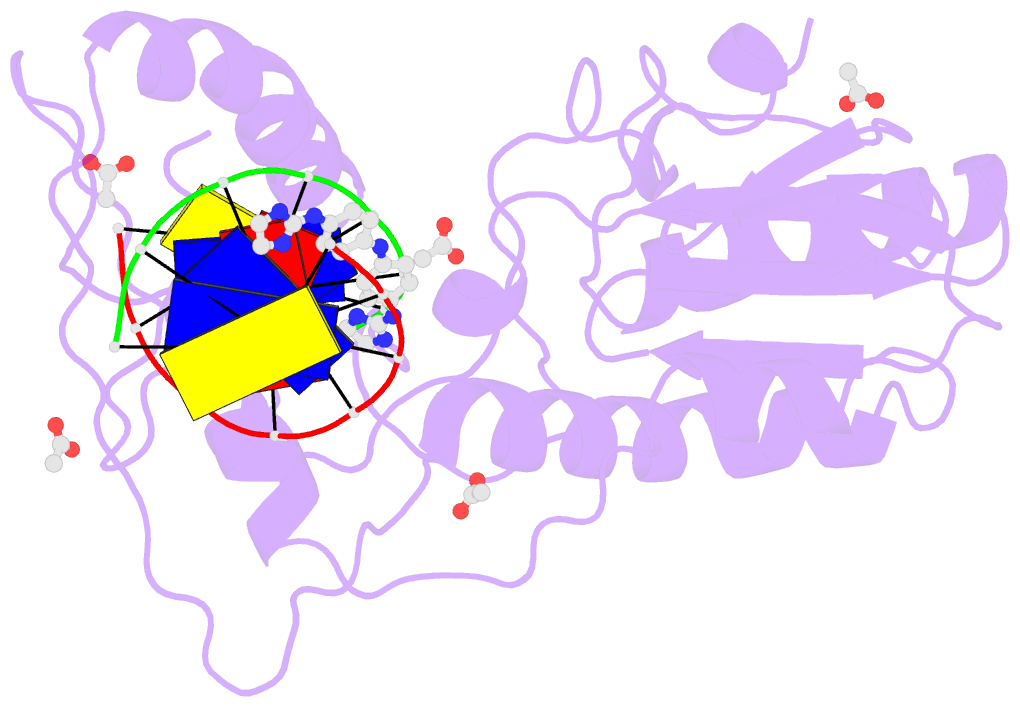
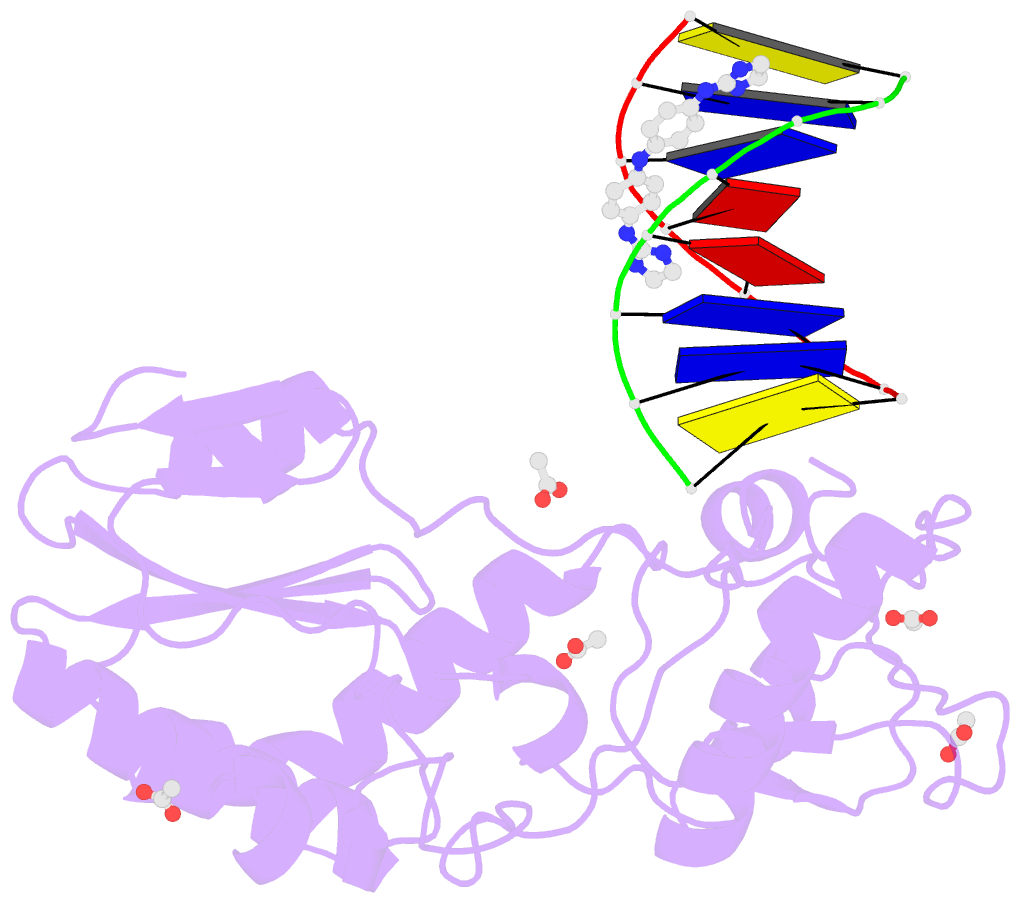
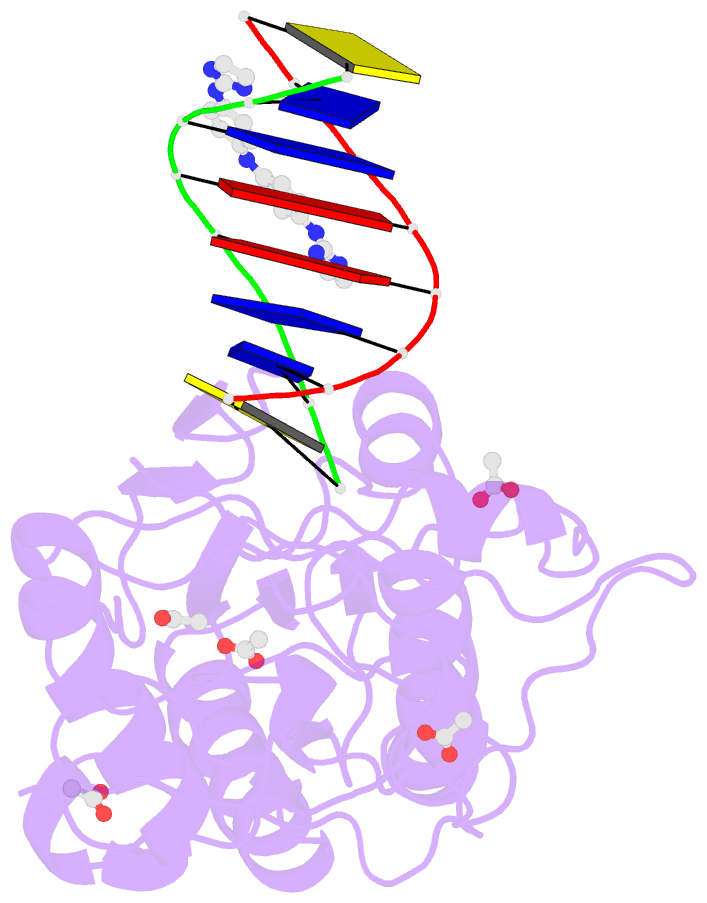
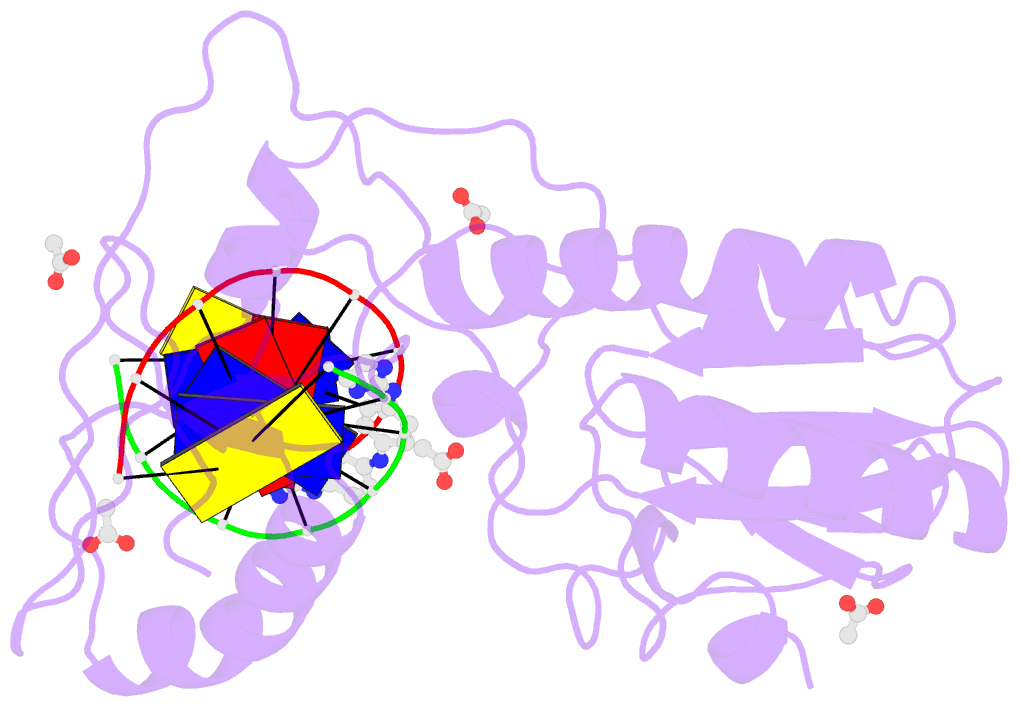
- Crystal structure of a trypanocidal 4,4'-bis(imidazolinylamino)diphenylamine bound to DNA
- Reference
- Glass LS, Nguyen B, Goodwin KD, Dardonville C, Wilson WD, Long EC, Georgiadis MM (2009): "Crystal structure of a trypanocidal 4,4'-bis(imidazolinylamino)diphenylamine bound to DNA." Biochemistry, 48, 5943-5952. doi: 10.1021/bi900204w.
- Abstract
- The pursuit of small molecules that bind to DNA has led to the discovery of selective and potent antitrypanosomal agents, specifically 4,4'-bis(imidazolinylamino)- and 4,4'-bis(guanidino)diphenylamine compounds, CD27 and CD25, respectively. Although the antitrypanosomal properties of these compounds have been characterized, further development of this series of compounds requires assessment of their DNA site selectivities and affinities. Toward this end, both compounds have been analyzed and found to selectively bind AT sequences. However, CD27 was found to bind with higher affinity to 5'-AATT than 5'-ATAT while CD25 bound more weakly but equally well to either sequence. To detail the nature of its interactions with DNA, the crystal structure of CD27, bound to its preferred DNA-binding site 5'-AATT within a self-complementary oligonucleotide, 5'-d(CTTAATTCGAATTAAG), was determined at 1.75 A using a host-guest approach. Although CD27 is predicted to be highly twisted in its energy-minimized state, it adopts a more planar crescent shape when bound in the minor groove of the DNA. Interactions of CD27 with 5'-AATT include bifurcated hydrogen bonds, providing a basis for selectivity of this site, and favorable van der Waals interactions in a slightly widened minor groove. Thus, an induced fit results from conformational changes in both the ligand and the DNA. Our studies suggest a basis for understanding the mechanism of the antitrypanosomal activity of these symmetric diphenylamine compounds.