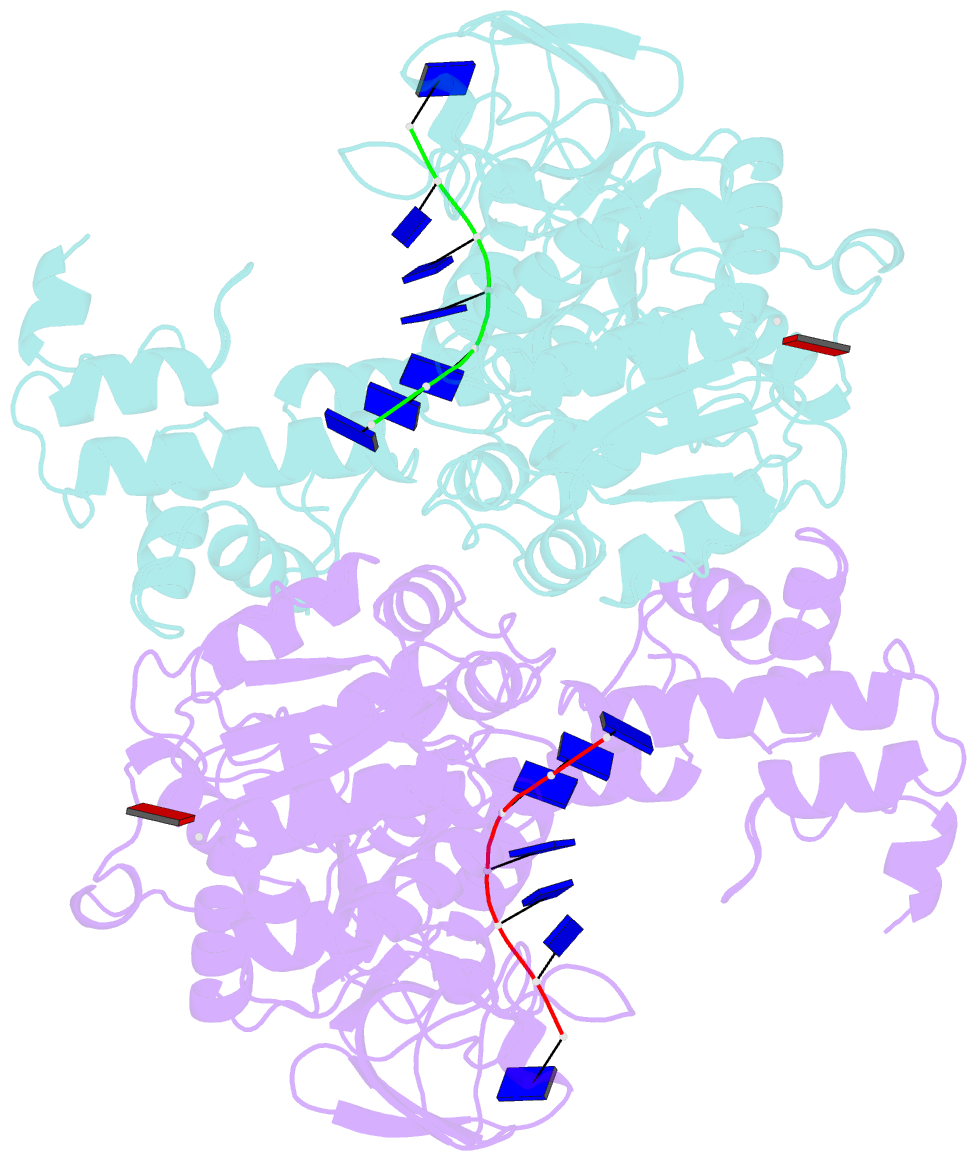
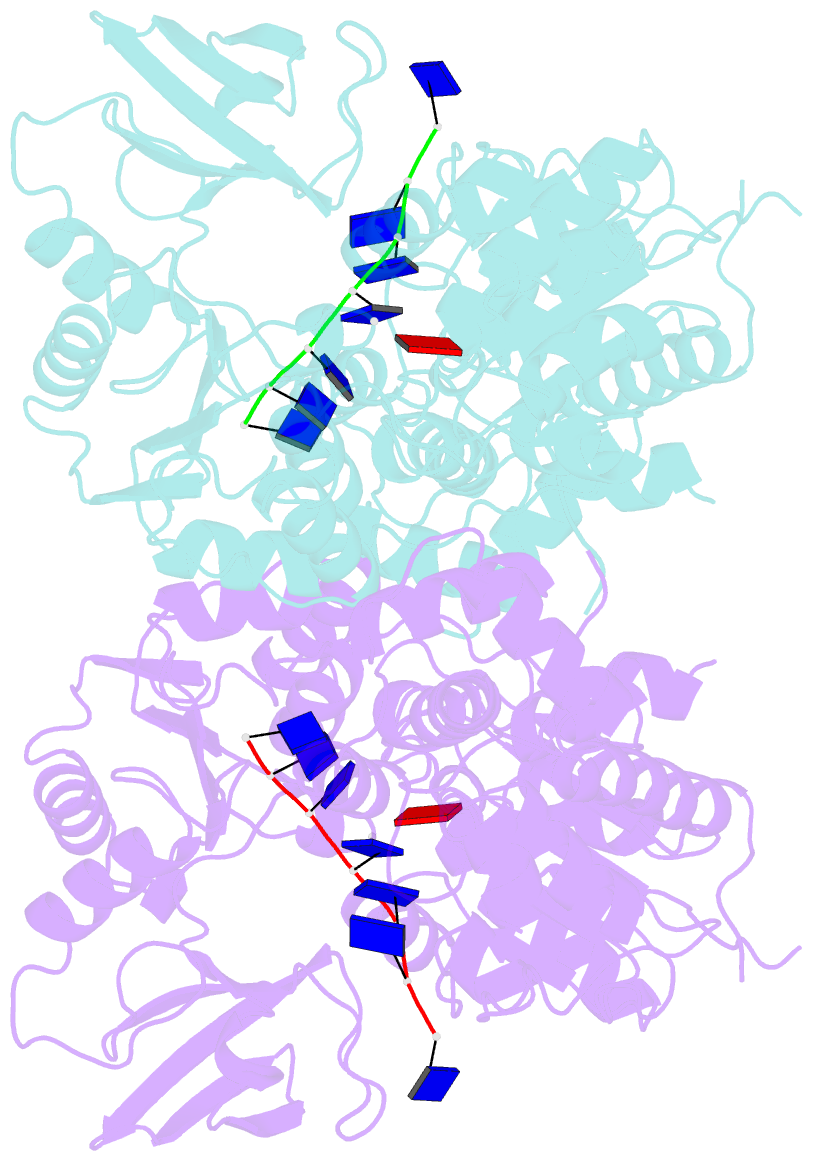
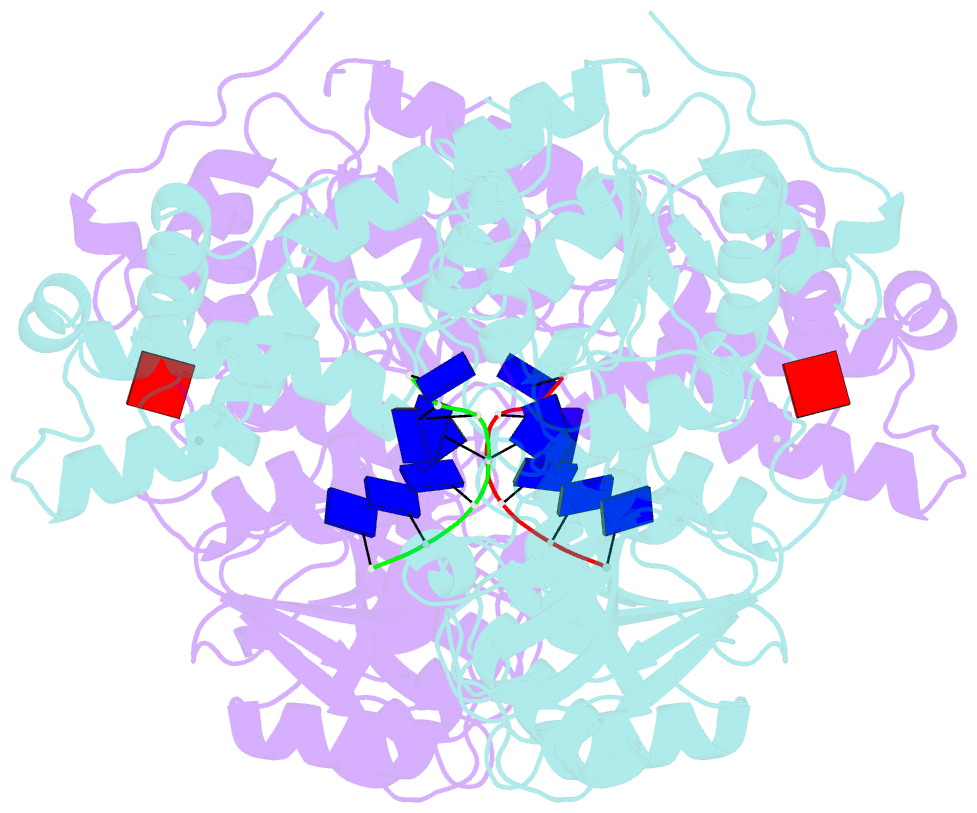
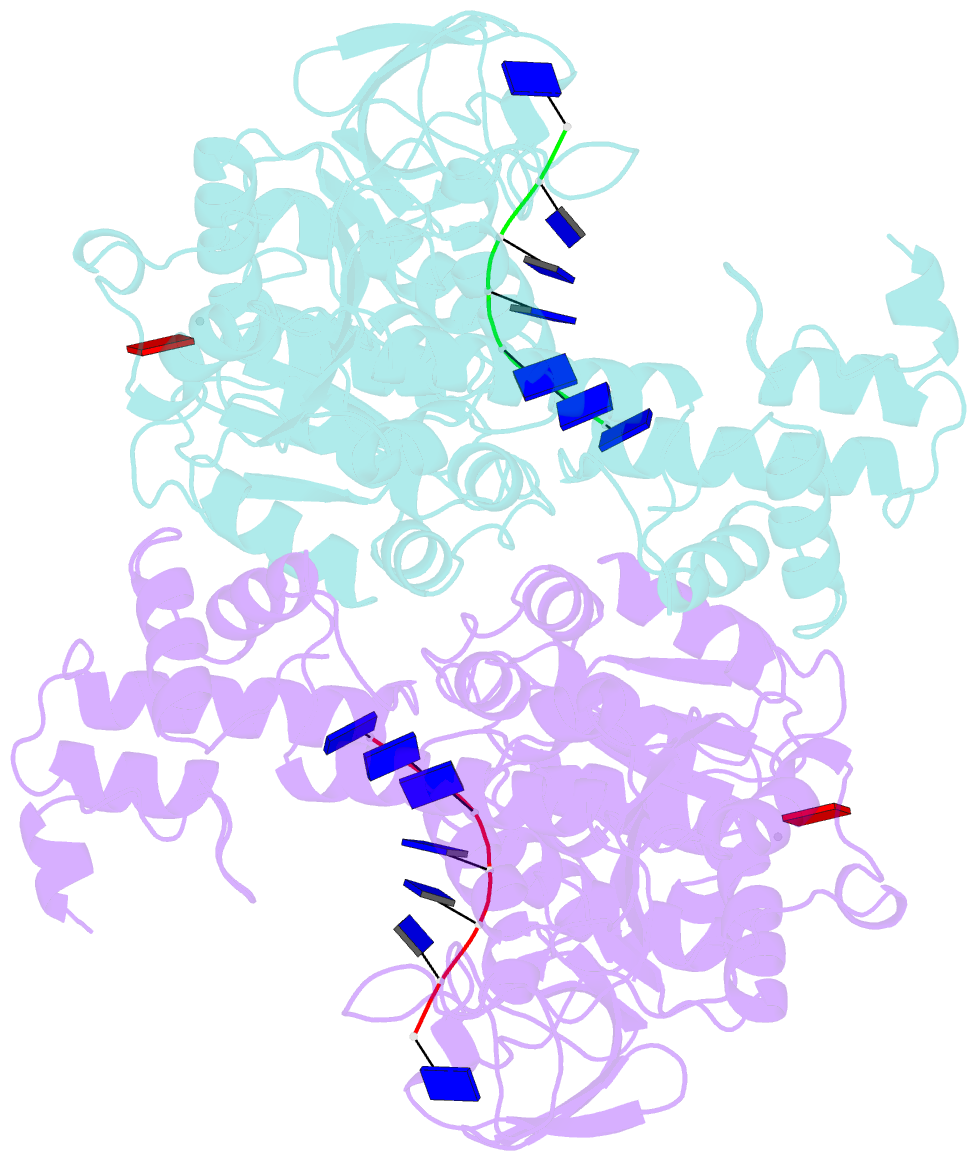
Summary information and primary citation
- PDB-id
- 3gpl; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.5 Å)
- Summary
- Crystal structure of the ternary complex of recd2 with DNA and adpnp
- Reference
- Saikrishnan K, Powell B, Cook NJ, Webb MR, Wigley DB (2009): "Mechanistic basis of 5'-3' translocation in SF1B helicases." Cell(Cambridge,Mass.), 137, 849-859. doi: 10.1016/j.cell.2009.03.036.
- Abstract
- Superfamily 1B (SF1B) helicases translocate in a 5'-3' direction and are required for a range of cellular activities across all domains of life. However, structural analyses to date have focused on how SF1A helicases achieve 3'-5' movement along nucleic acids. We present crystal structures of the complex between the SF1B helicase RecD2 from Deinococcus radiodurans and ssDNA in the presence and absence of an ATP analog. These snapshots of the reaction pathway reveal a nucleotide binding-induced conformational change of the two motor domains that is broadly reminiscent of changes observed in other SF1 and SF2 helicases. Together with biochemical data, the structures point to a step size for translocation of one base per ATP hydrolyzed. Moreover, the structures also reveal a mechanism for nucleic acid translocation in the 5'-3' direction by SF1B helicases that is surprisingly different from that of 3'-5' translocation by SF1A enzymes, and explains the molecular basis of directionality.