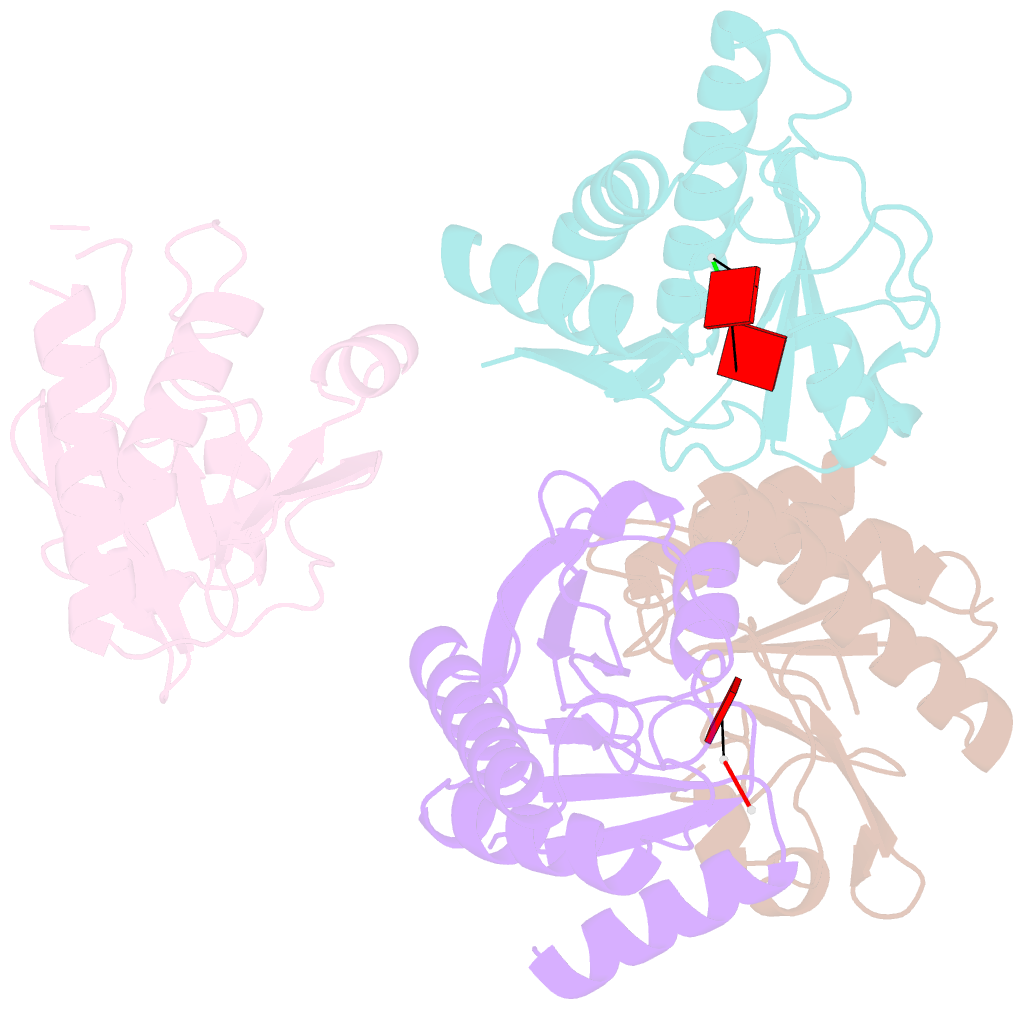
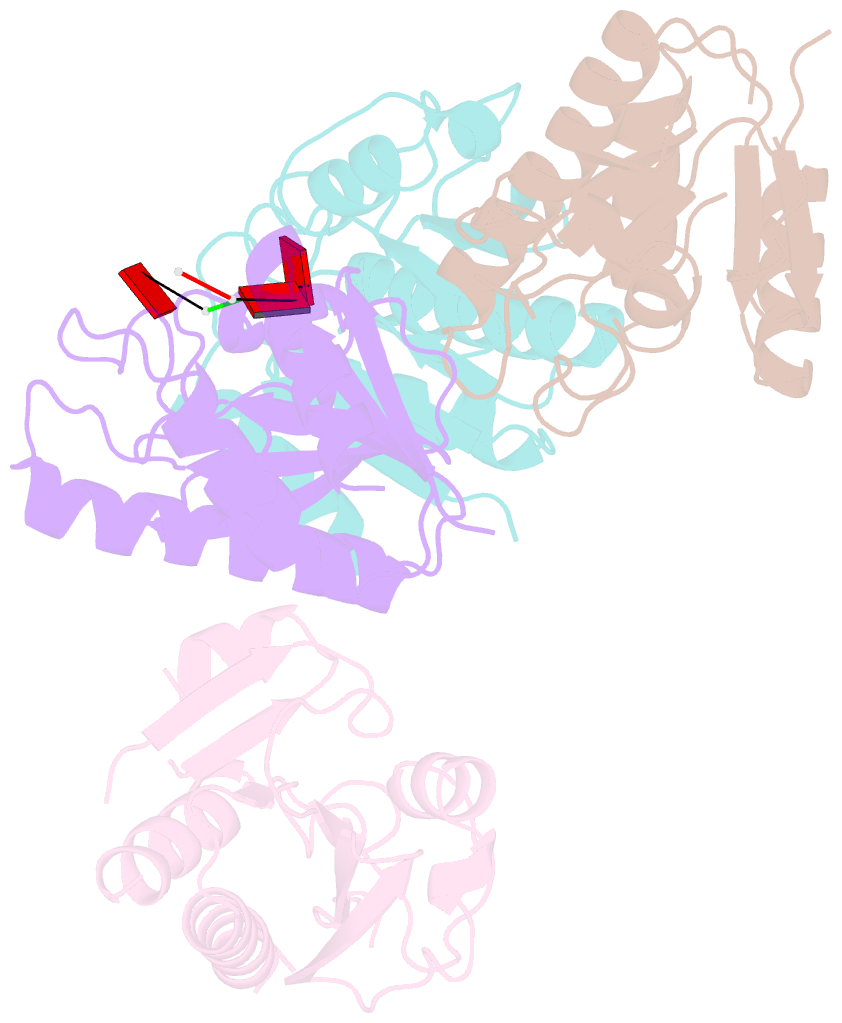
Summary information and primary citation
- PDB-id
- 3gpq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- X-ray (2.0 Å)
- Summary
- Crystal structure of macro domain of chikungunya virus in complex with RNA
- Reference
- Malet H, Coutard B, Jamal S, Dutartre H, Papageorgiou N, Neuvonen M, Ahola T, Forrester N, Gould EA, Lafitte D, Ferron F, Lescar J, Gorbalenya AE, de Lamballerie X, Canard B (2009): "The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket." J.Virol., 83, 6534-6545. doi: 10.1128/JVI.00189-09.
- Abstract
- Macro domains (also called "X domains") constitute a protein module family present in all kingdoms of life, including viruses of the Coronaviridae and Togaviridae families. Crystal structures of the macro domain from the Chikungunya virus (an "Old World" alphavirus) and the Venezuelan equine encephalitis virus (a "New World" alphavirus) were determined at resolutions of 1.65 and 2.30 A, respectively. These domains are active as adenosine di-phosphoribose 1''-phosphate phosphatases. Both the Chikungunya and the Venezuelan equine encephalitis virus macro domains are ADP-ribose binding modules, as revealed by structural and functional analysis. A single aspartic acid conserved through all macro domains is responsible for the specific binding of the adenine base. Sequence-unspecific binding to long, negatively charged polymers such as poly(ADP-ribose), DNA, and RNA is observed and attributed to positively charged patches outside of the active site pocket, as judged by mutagenesis and binding studies. The crystal structure of the Chikungunya virus macro domain with an RNA trimer shows a binding mode utilizing the same adenine-binding pocket as ADP-ribose, but avoiding the ADP-ribose 1''-phosphate phosphatase active site. This leaves the AMP binding site as the sole common feature in all macro domains.