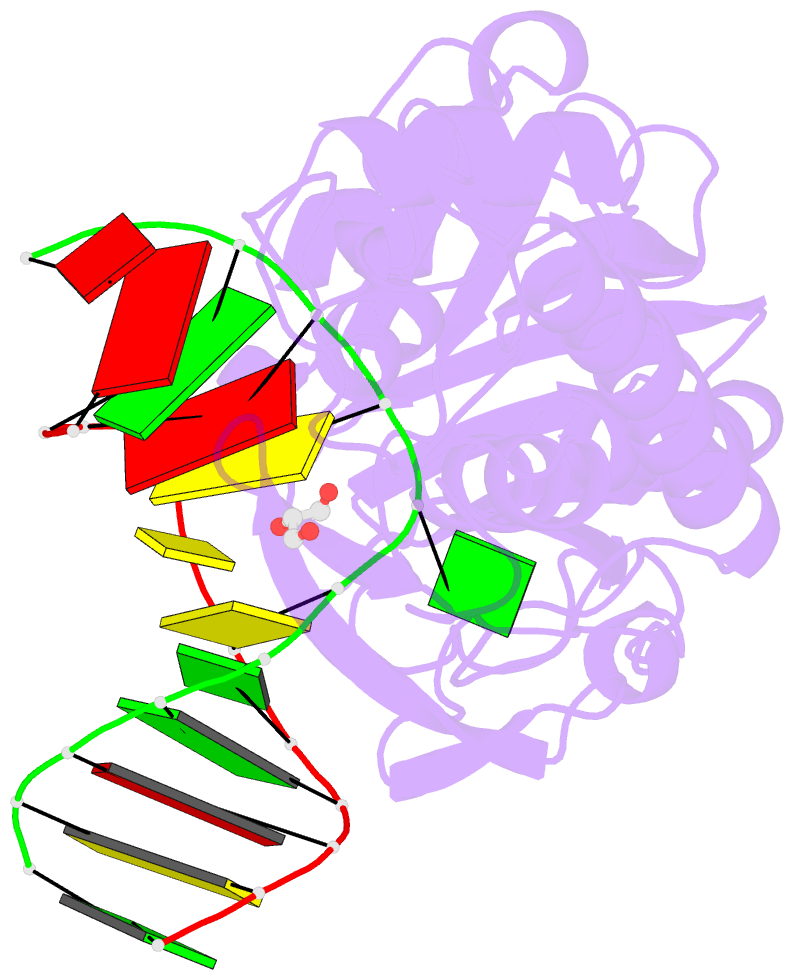
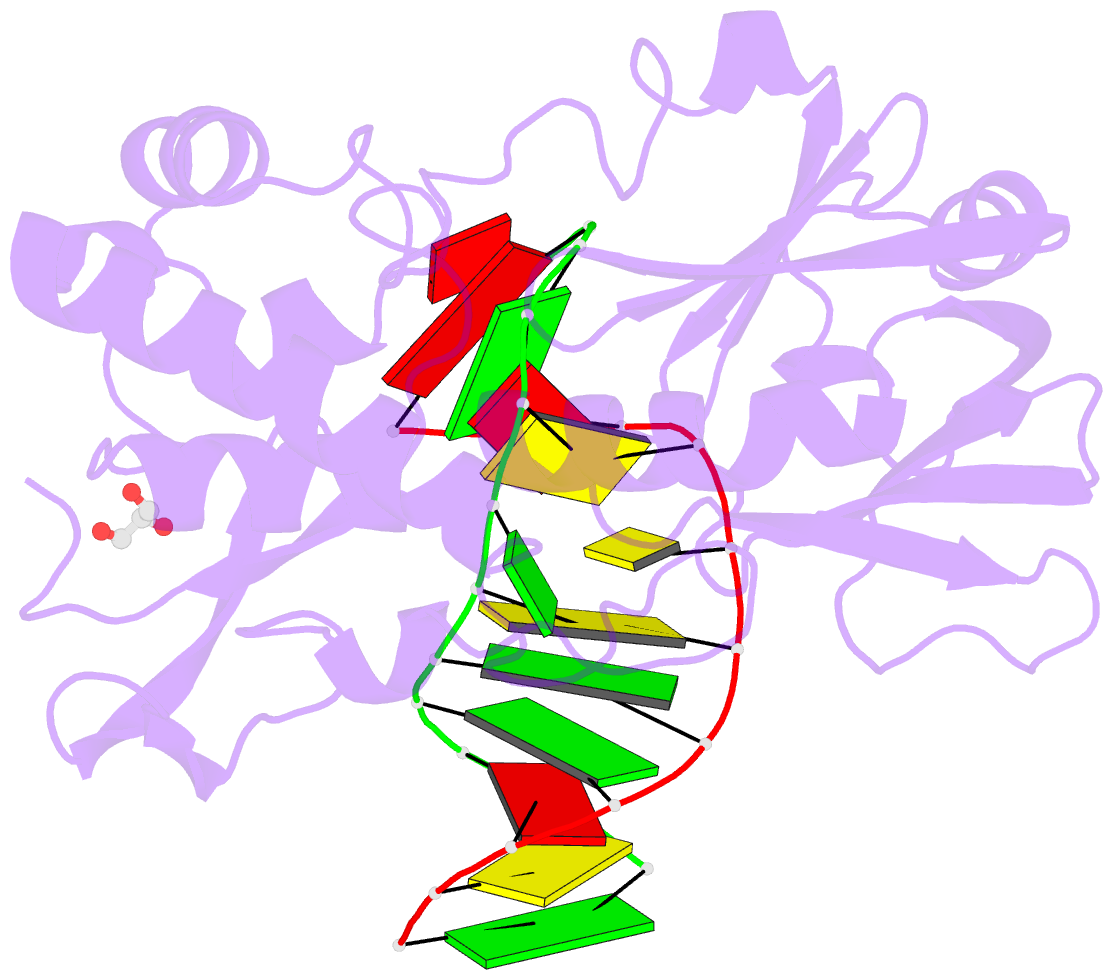
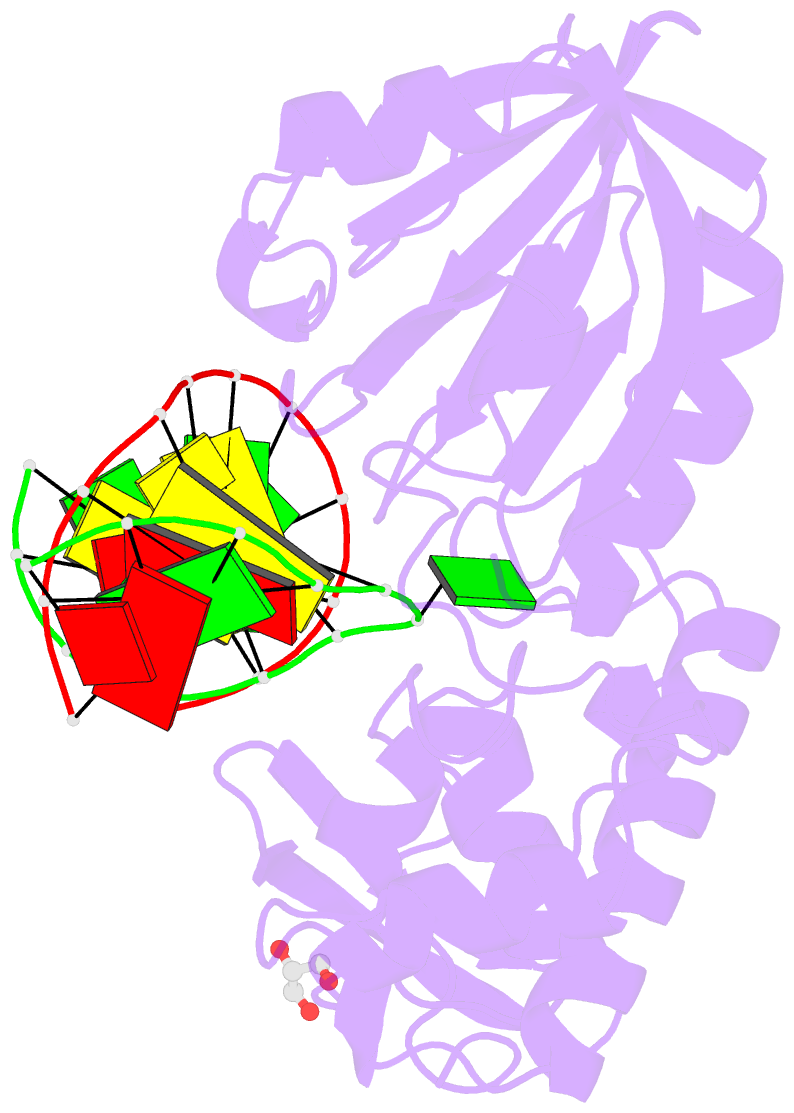
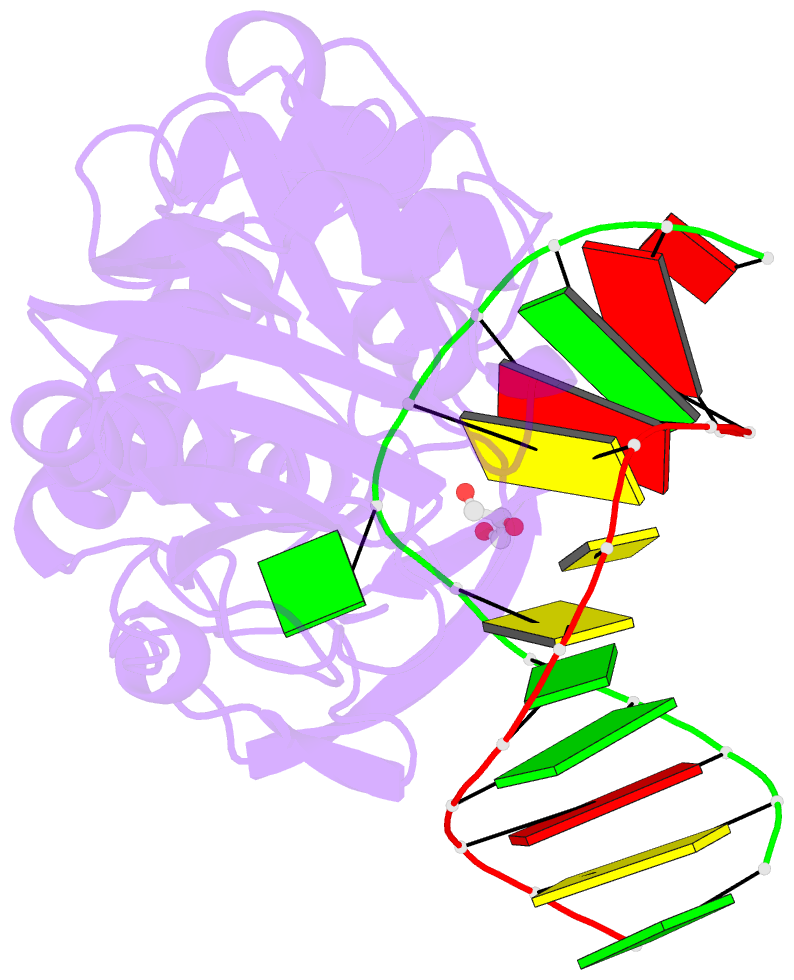
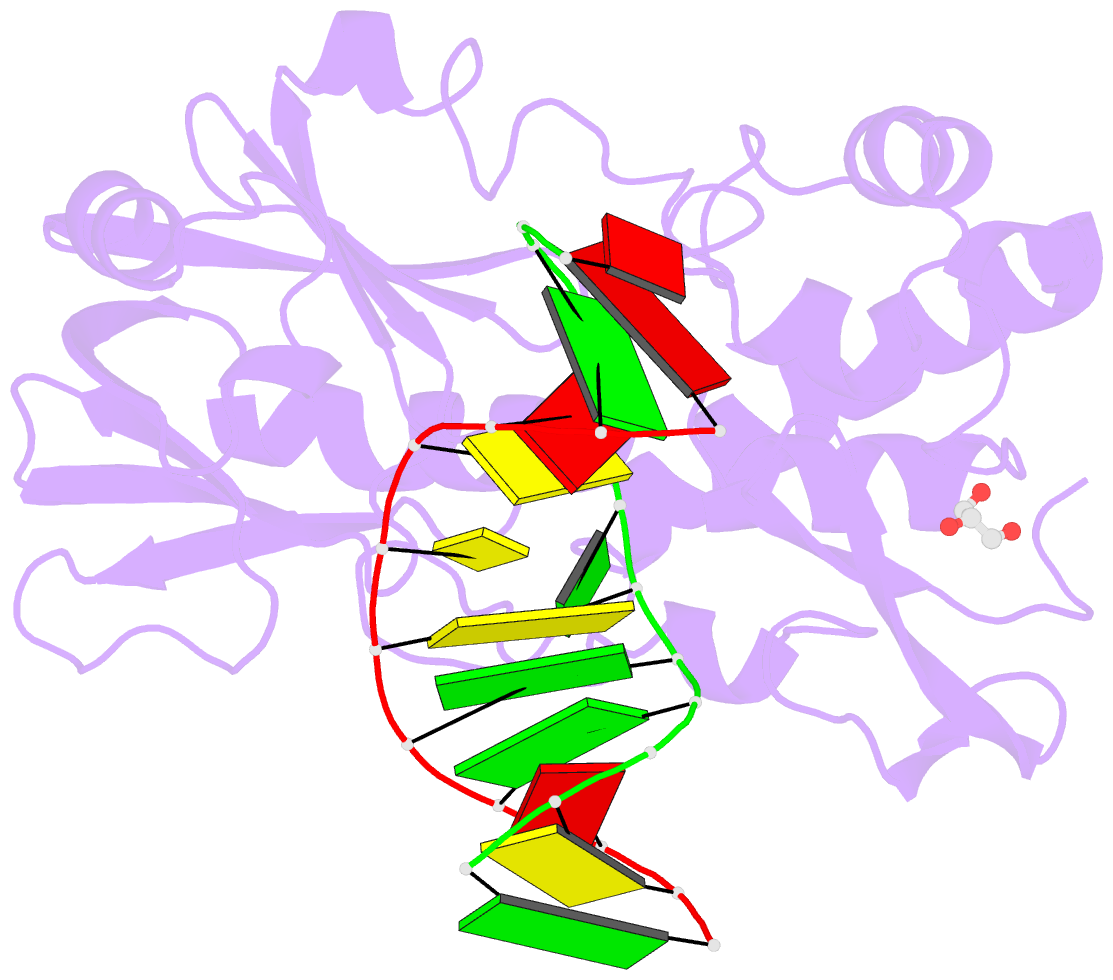
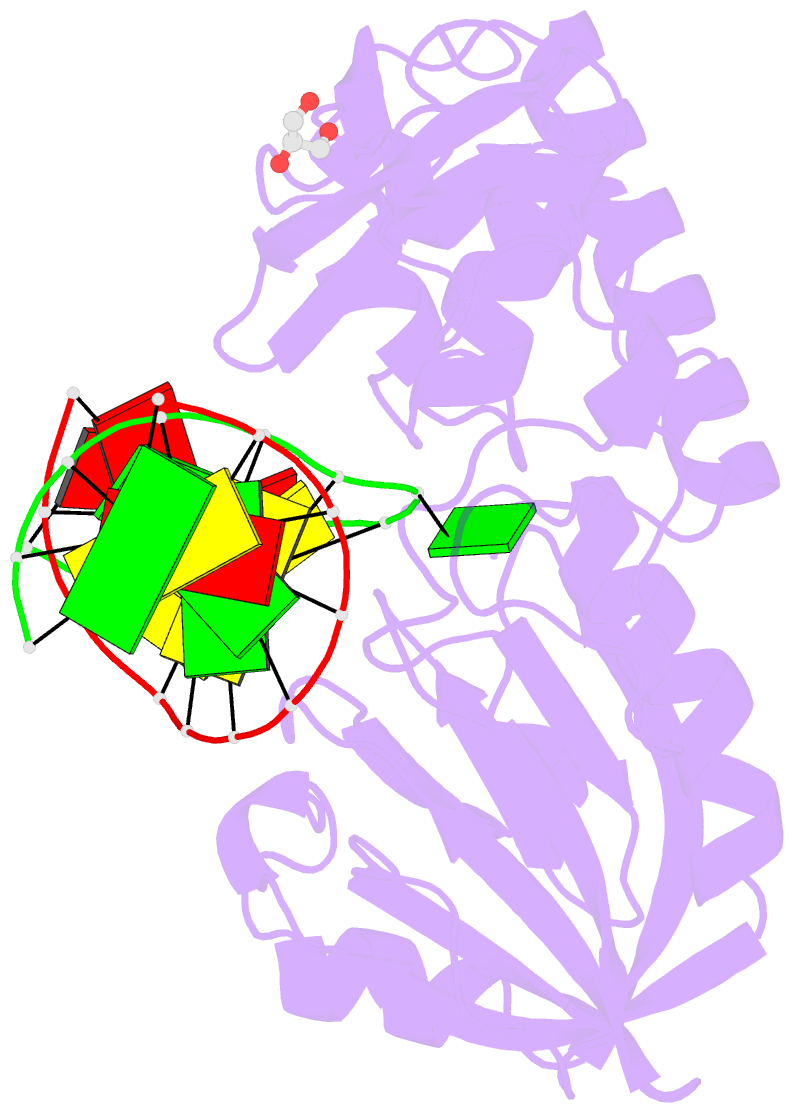
Summary information and primary citation
- PDB-id
- 3gq4; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- lyase-DNA
- Method
- X-ray (1.7 Å)
- Summary
- Sequence-matched mutm lesion recognition complex 5 (lrc5)
- Reference
- Qi Y, Spong MC, Nam K, Banerjee A, Jiralerspong S, Karplus M, Verdine GL (2009): "Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme." Nature, 462, 762-766. doi: 10.1038/nature08561.
- Abstract
- How living systems detect the presence of genotoxic damage embedded in a million-fold excess of undamaged DNA is an unresolved question in biology. Here we have captured and structurally elucidated a base-excision DNA repair enzyme, MutM, at the stage of initial encounter with a damaged nucleobase, 8-oxoguanine (oxoG), nested within a DNA duplex. Three structures of intrahelical oxoG-encounter complexes are compared with sequence-matched structures containing a normal G base in place of an oxoG lesion. Although the protein-DNA interfaces in the matched complexes differ by only two atoms-those that distinguish oxoG from G-their pronounced structural differences indicate that MutM can detect a lesion in DNA even at the earliest stages of encounter. All-atom computer simulations show the pathway by which encounter of the enzyme with the lesion causes extrusion from the DNA duplex, and they elucidate the critical free energy difference between oxoG and G along the extrusion pathway.