Summary information and primary citation
- PDB-id
- 3gtj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription, transferase-DNA-RNA hybrid
- Method
- X-ray (3.42 Å)
- Summary
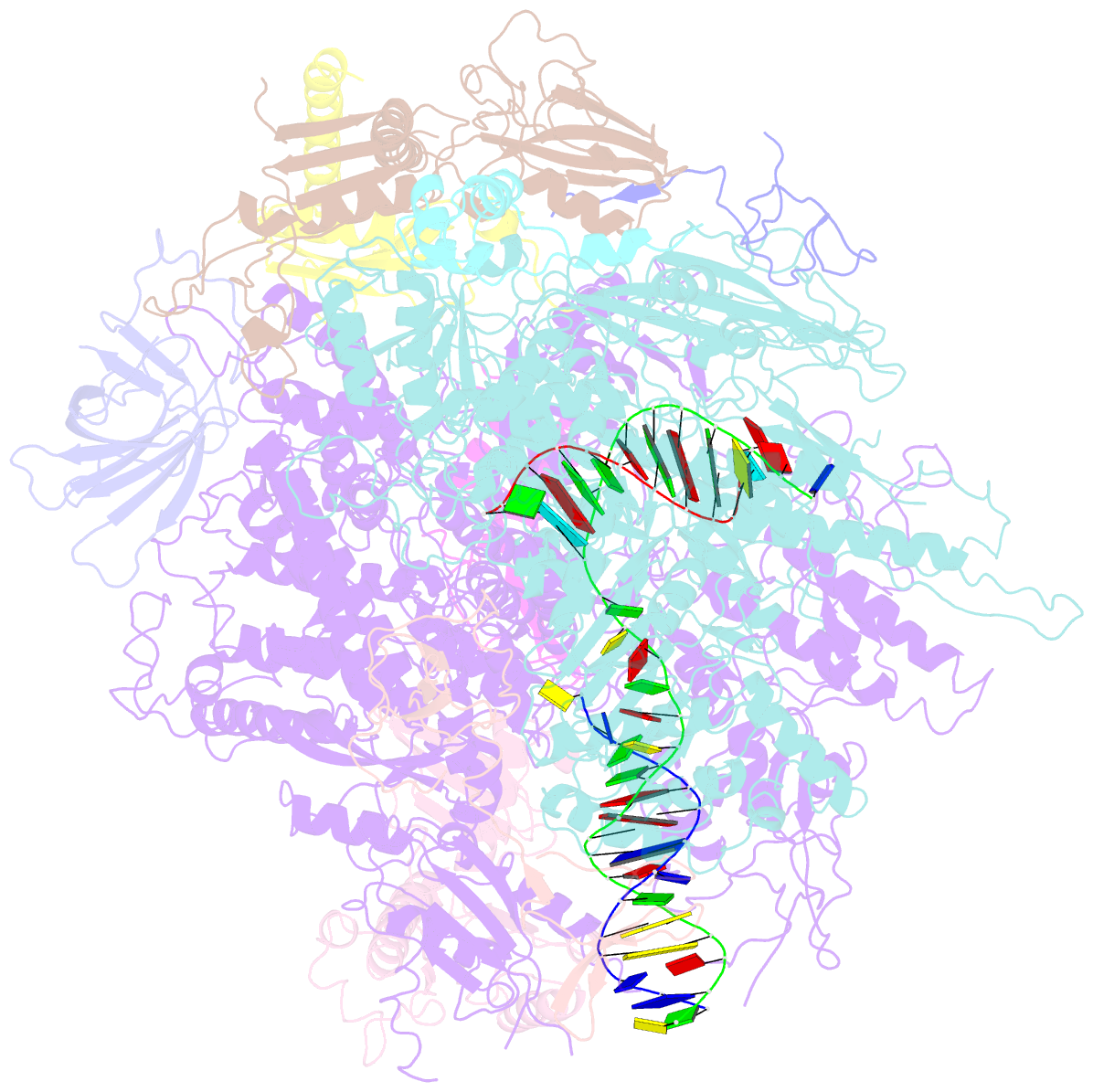
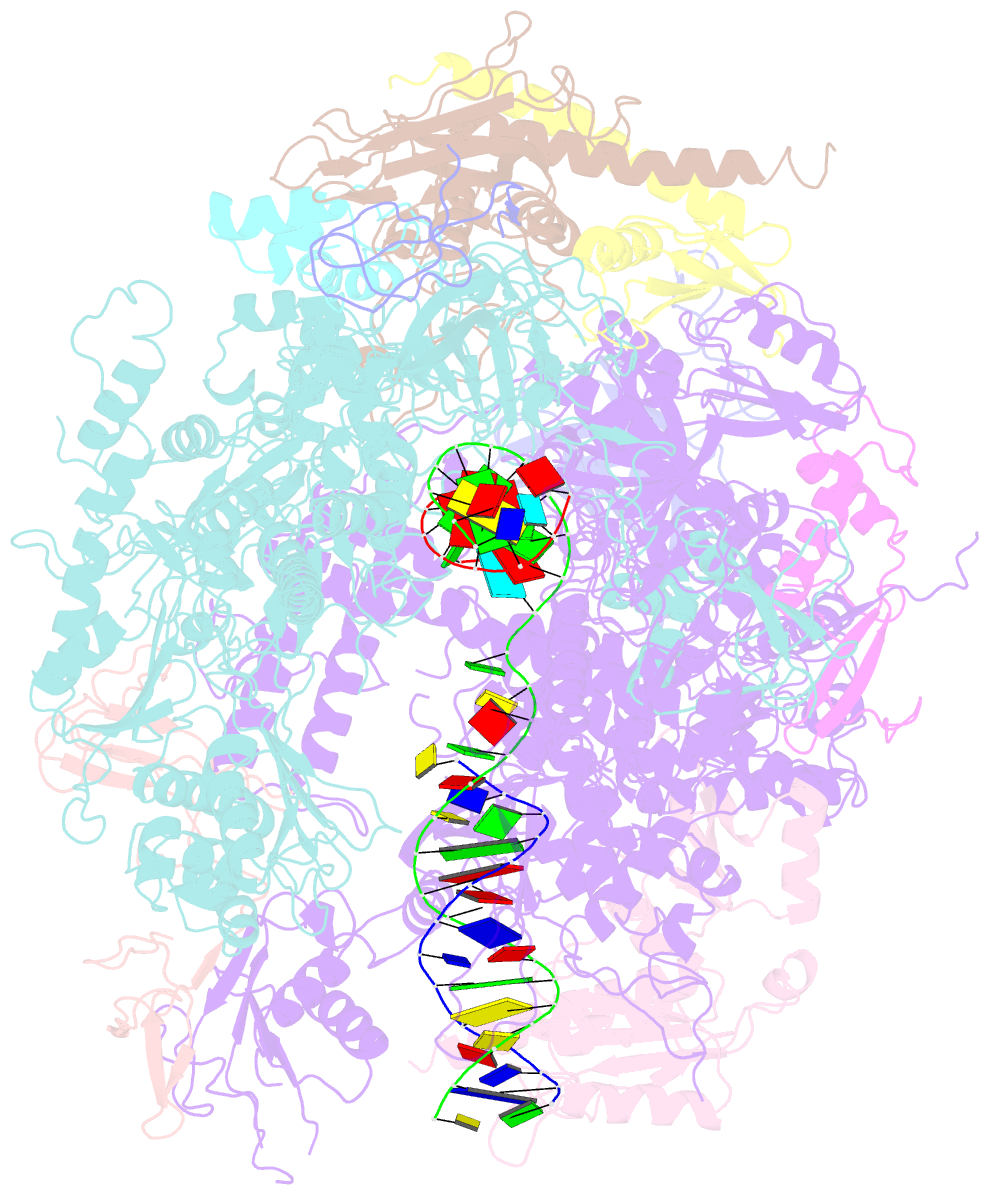
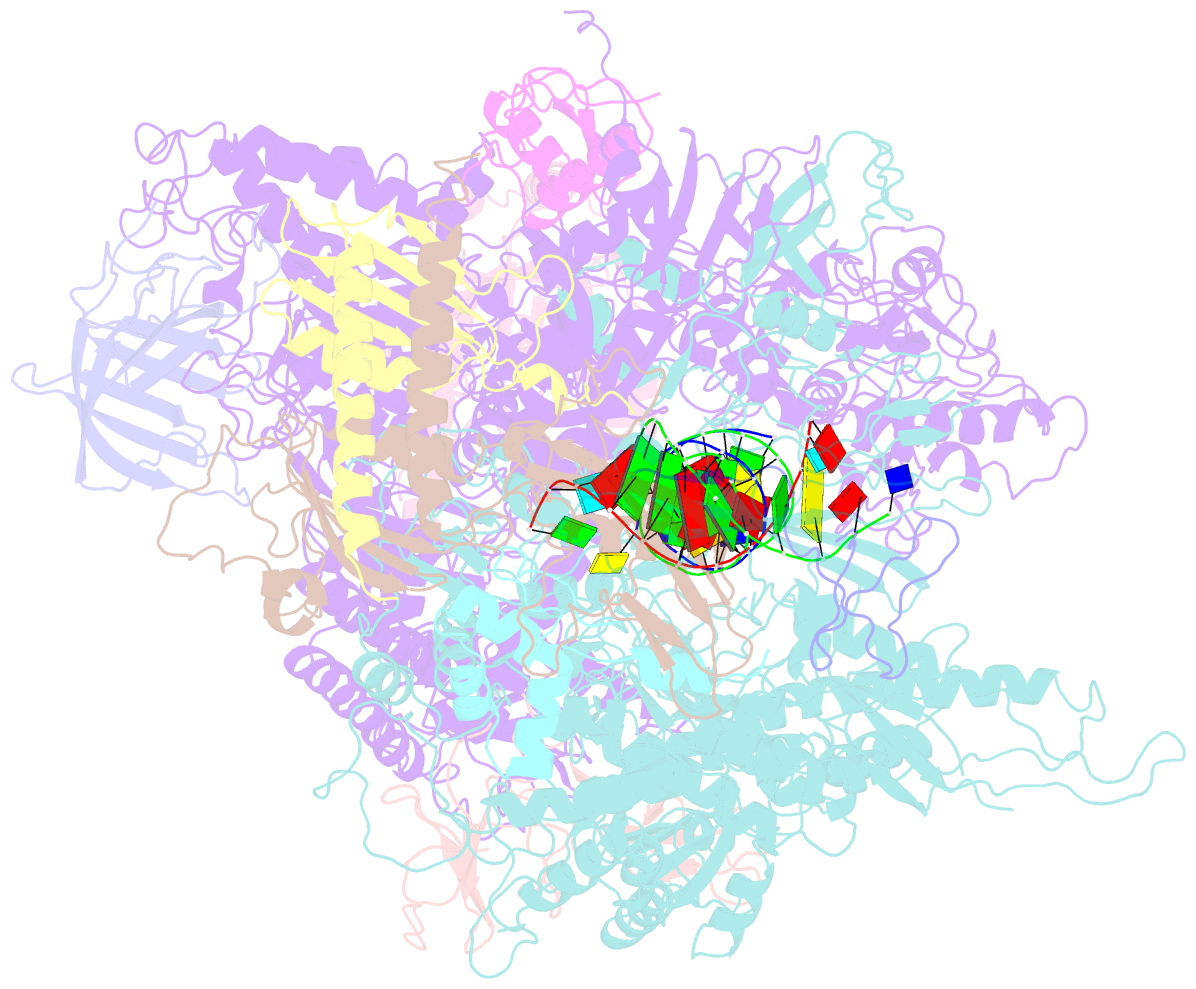
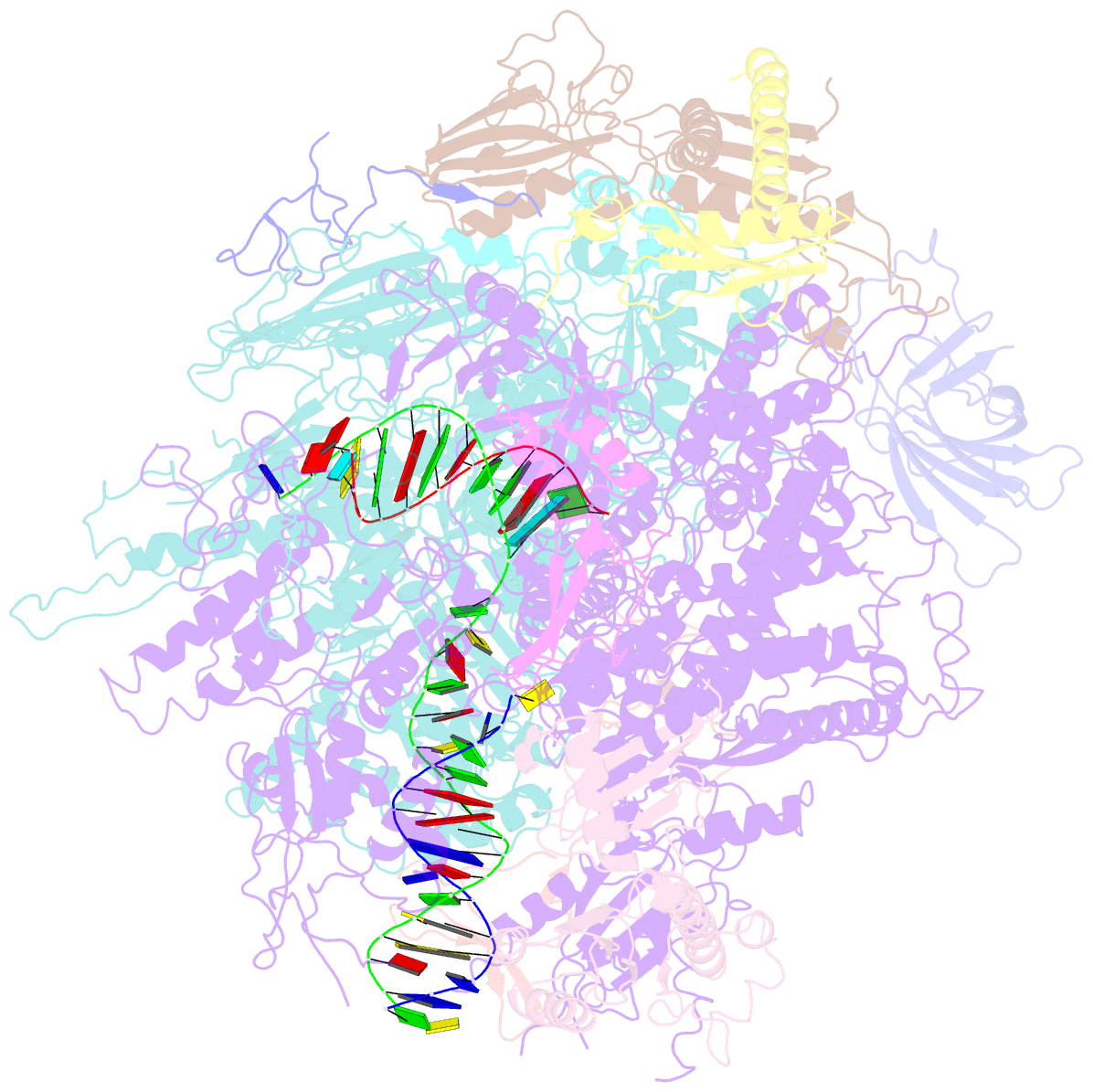
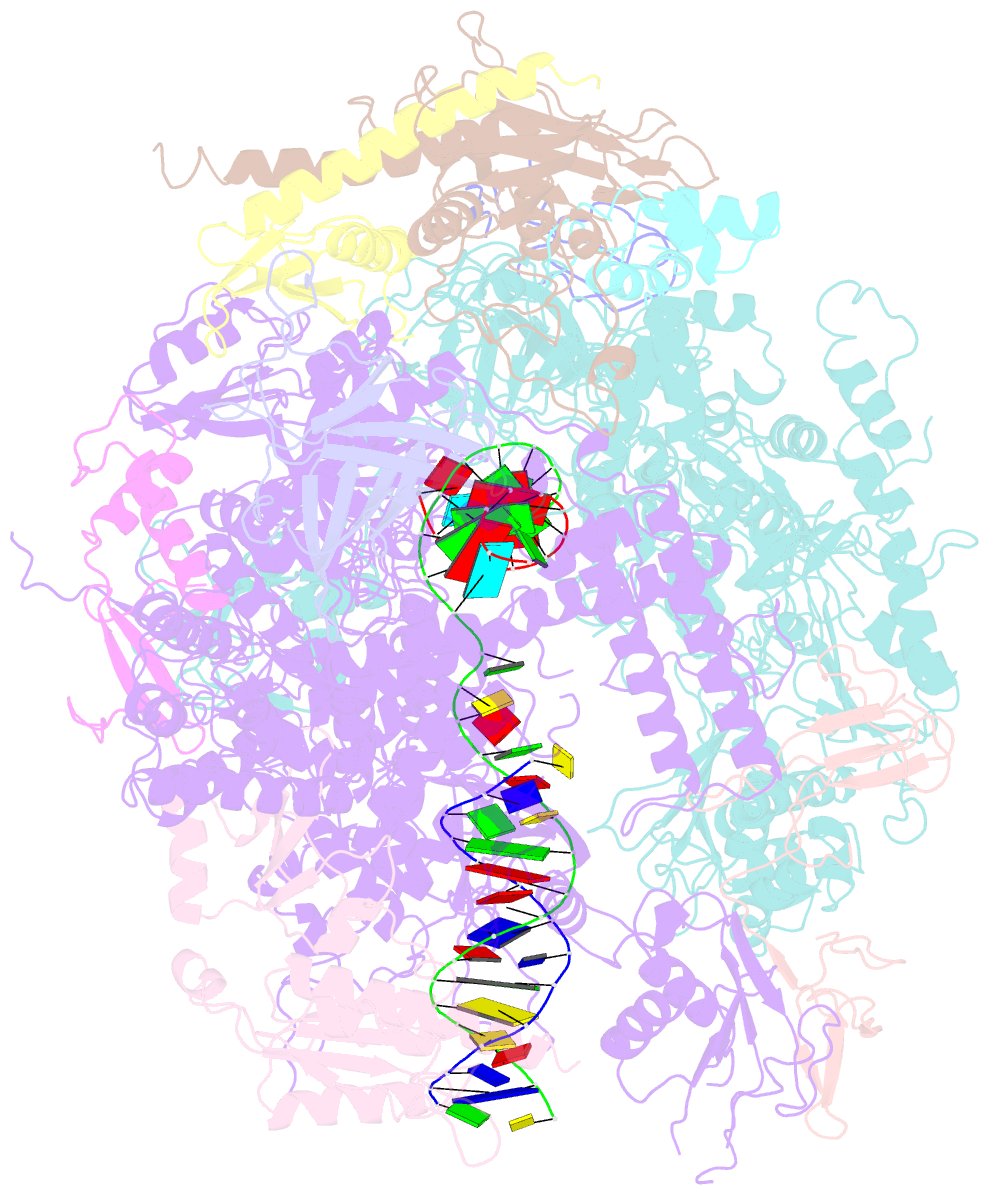
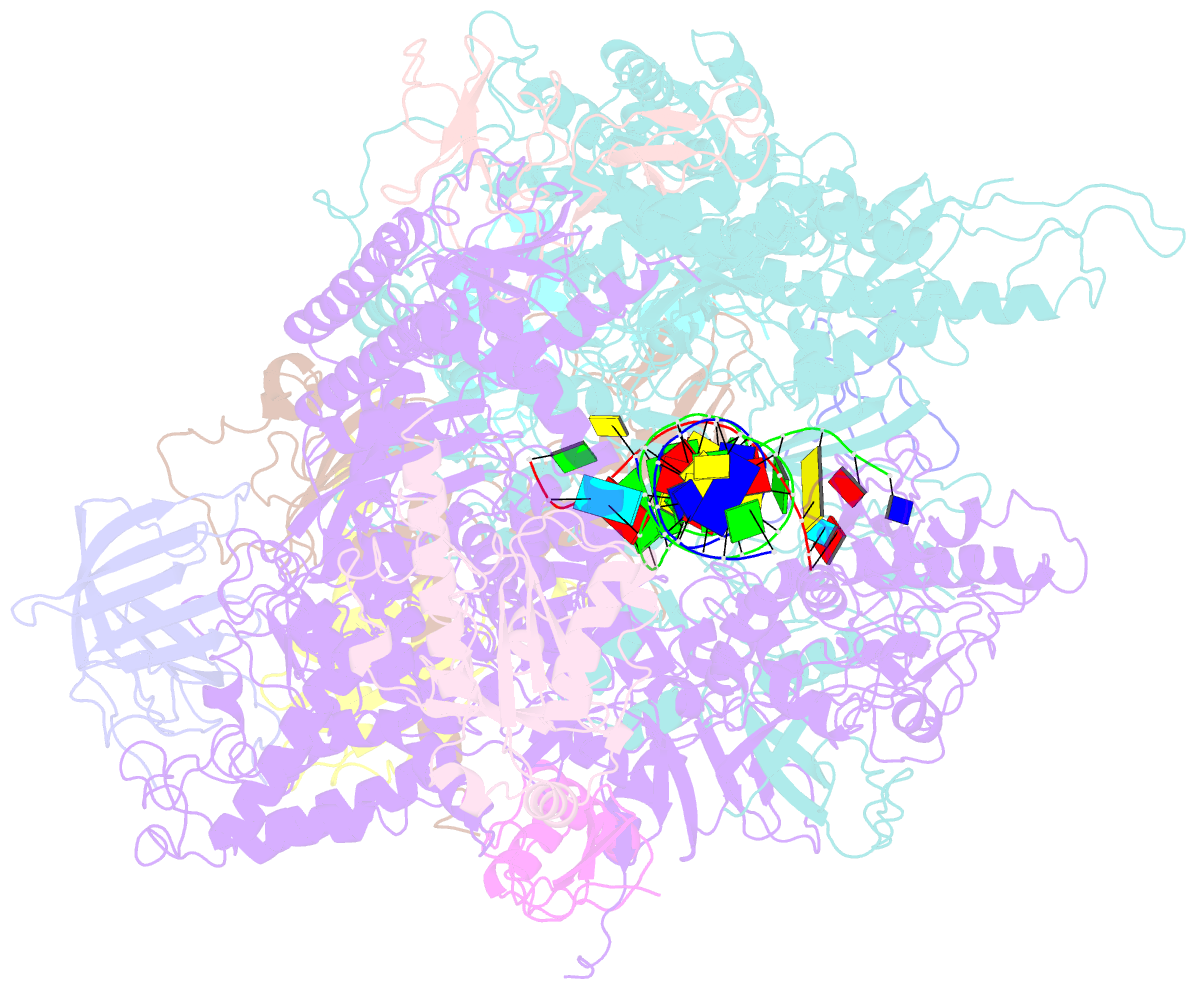
- Backtracked RNA polymerase ii complex with 13mer RNA
- Reference
- Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD (2009): "Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution." Science, 324, 1203-1206. doi: 10.1126/science.1168729.
- Abstract
- Transcribing RNA polymerases oscillate between three stable states, two of which, pre- and posttranslocated, were previously subjected to x-ray crystal structure determination. We report here the crystal structure of RNA polymerase II in the third state, the reverse translocated, or "backtracked" state. The defining feature of the backtracked structure is a binding site for the first backtracked nucleotide. This binding site is occupied in case of nucleotide misincorporation in the RNA or damage to the DNA, and is termed the "P" site because it supports proofreading. The predominant mechanism of proofreading is the excision of a dinucleotide in the presence of the elongation factor SII (TFIIS). Structure determination of a cocrystal with TFIIS reveals a rearrangement whereby cleavage of the RNA may take place.