Summary information and primary citation
- PDB-id
- 3hhz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- X-ray (3.5 Å)
- Summary
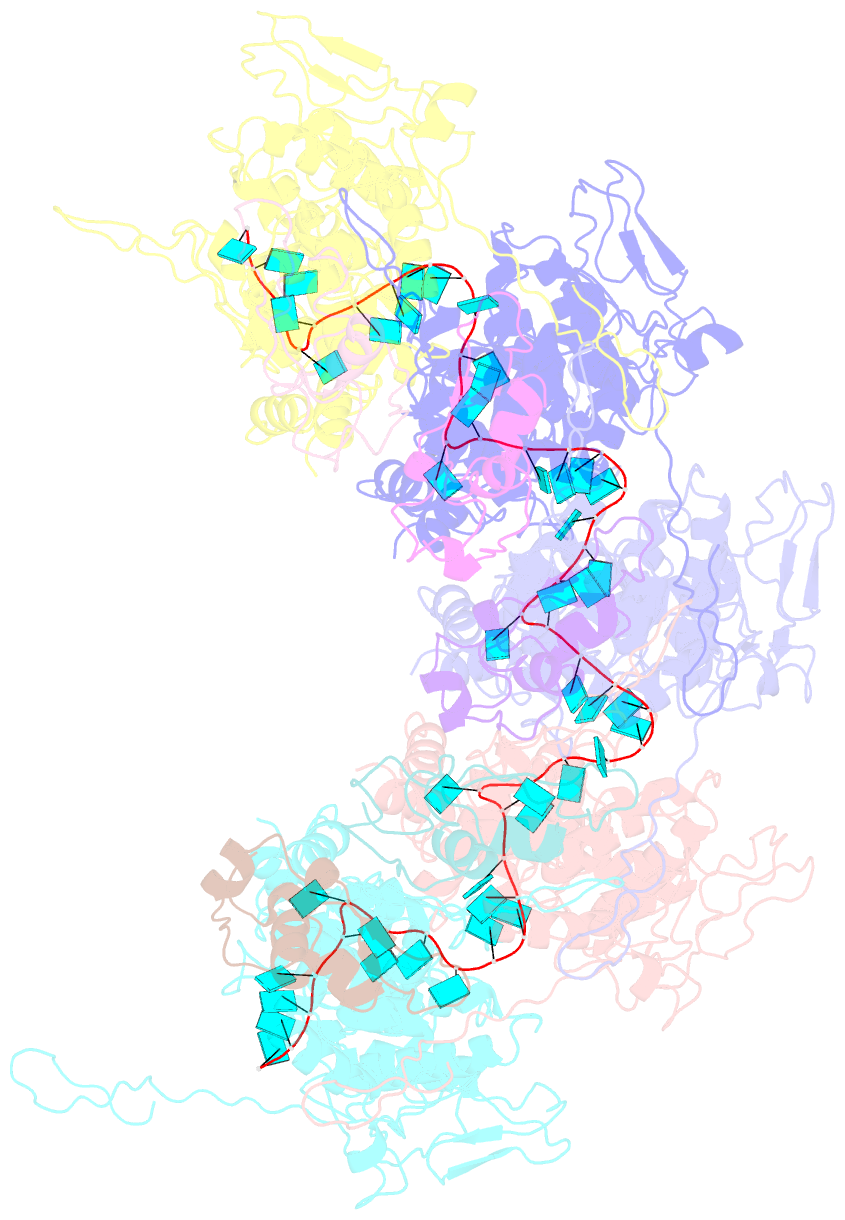
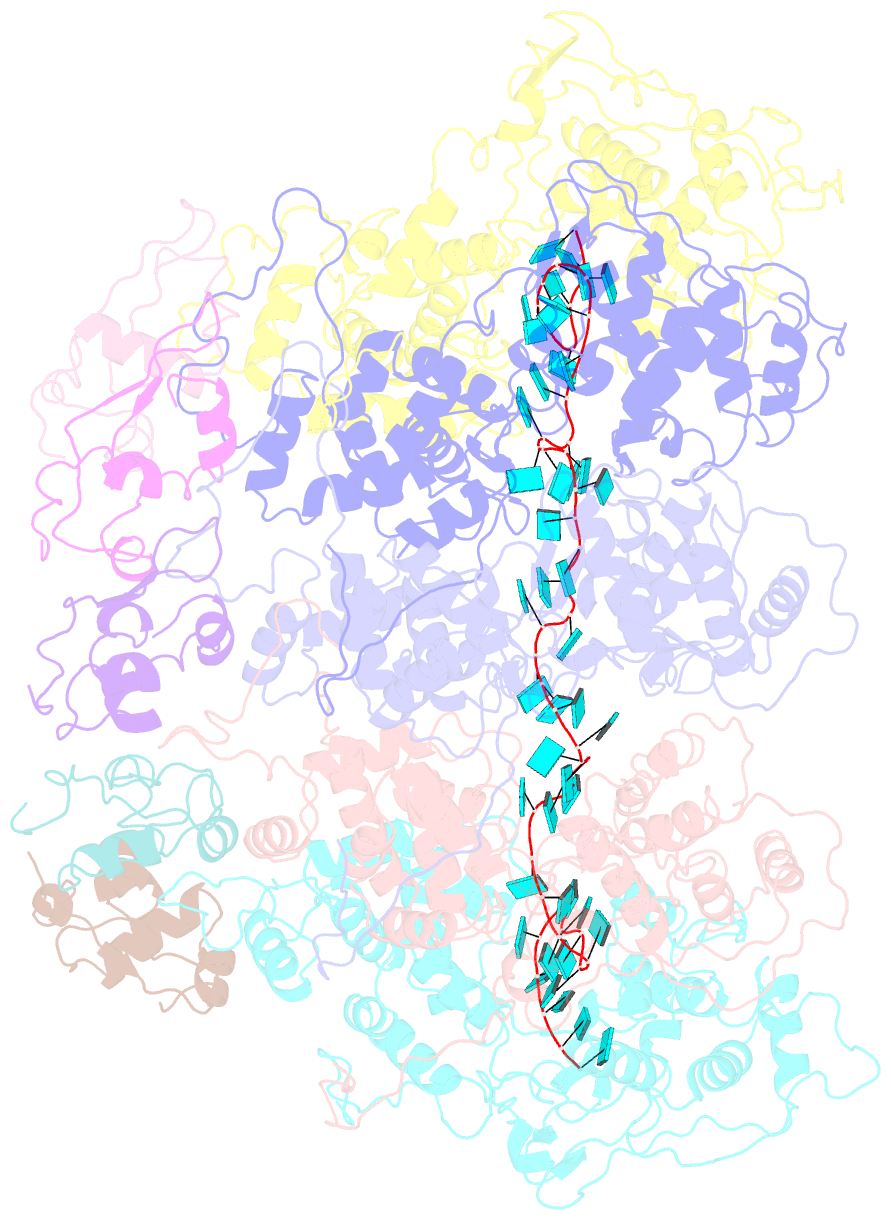
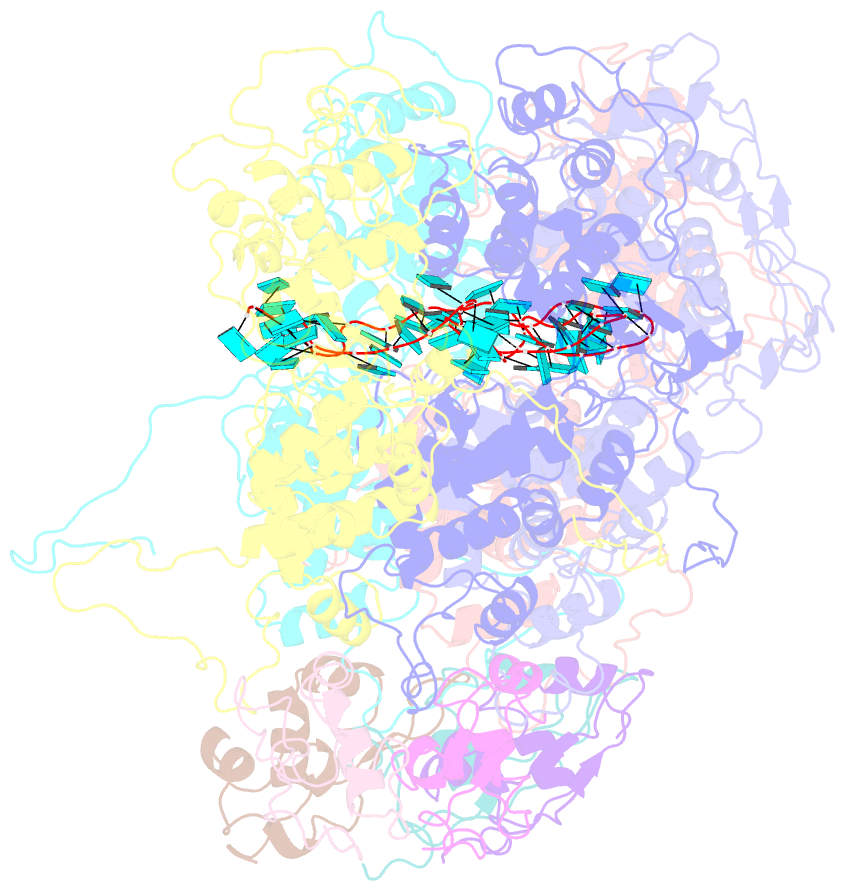
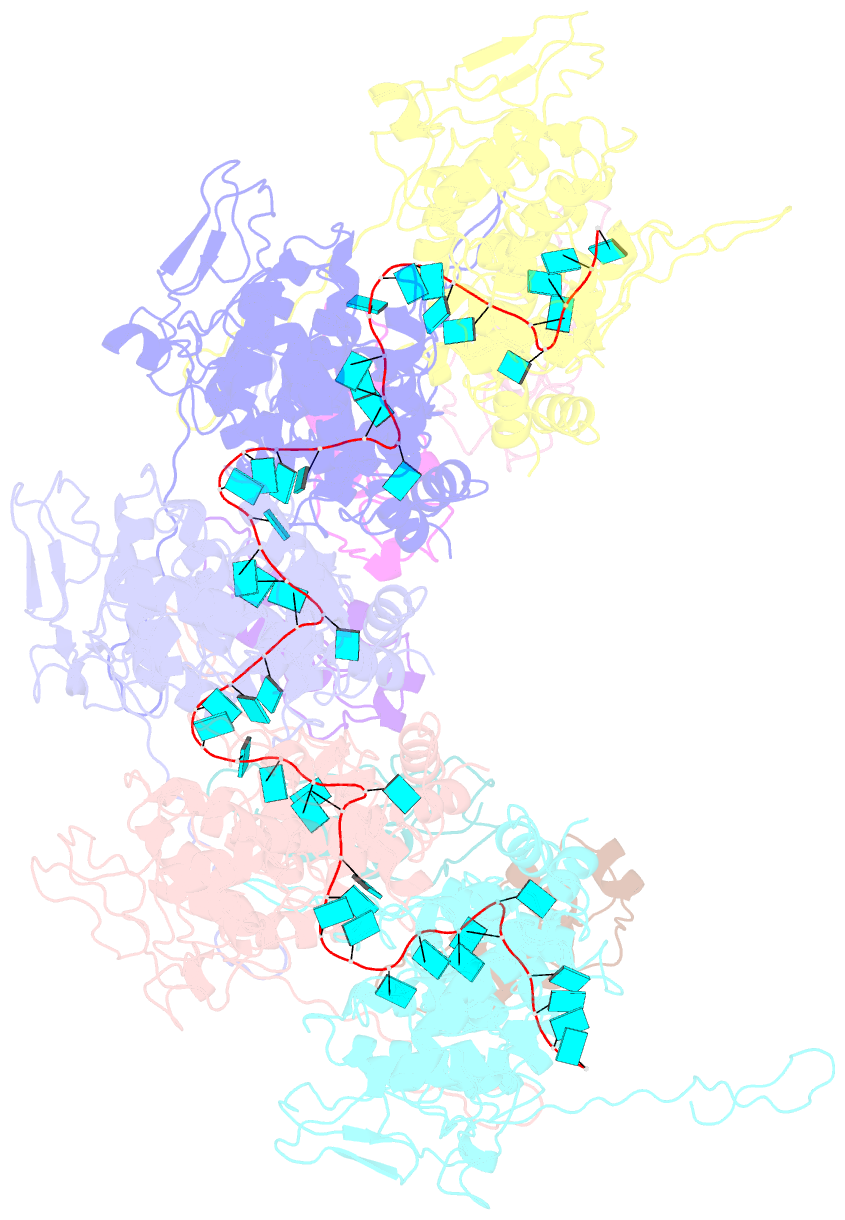
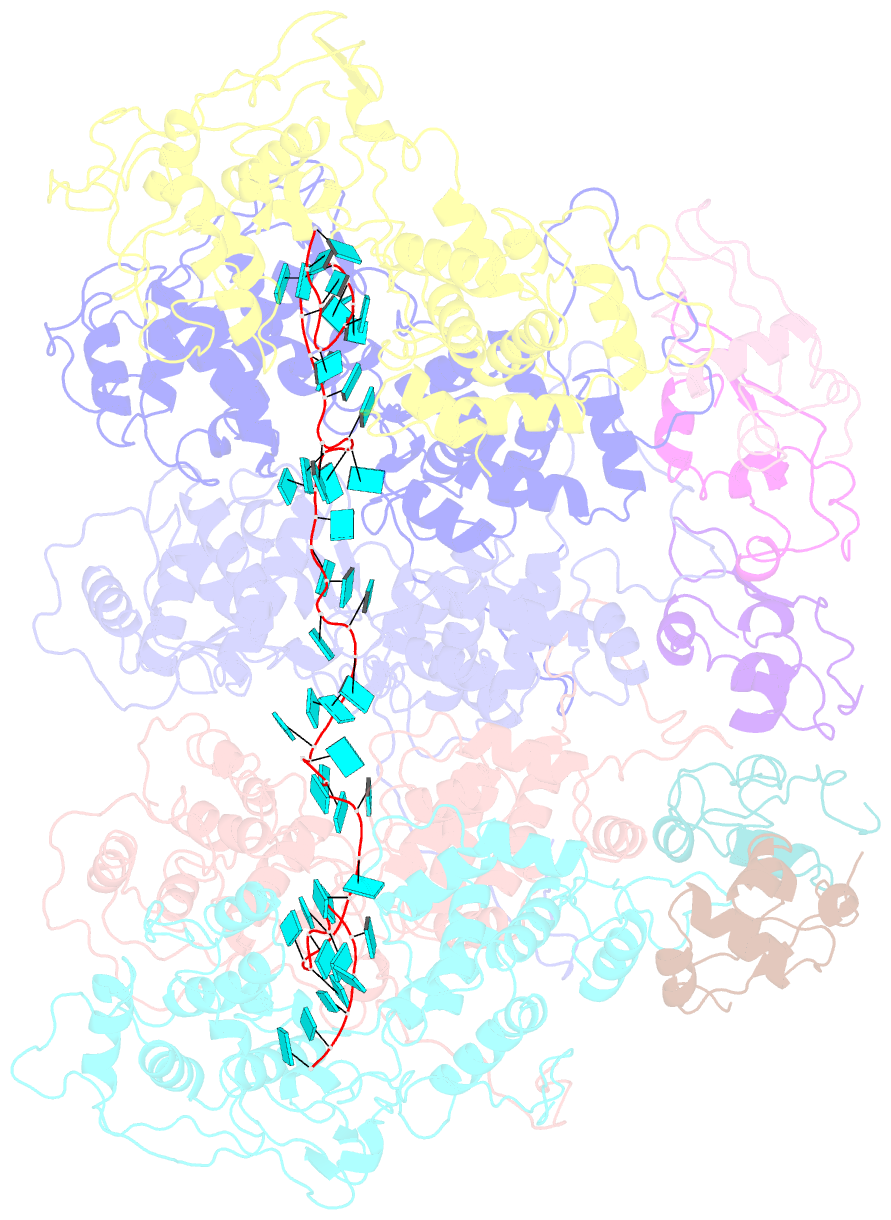
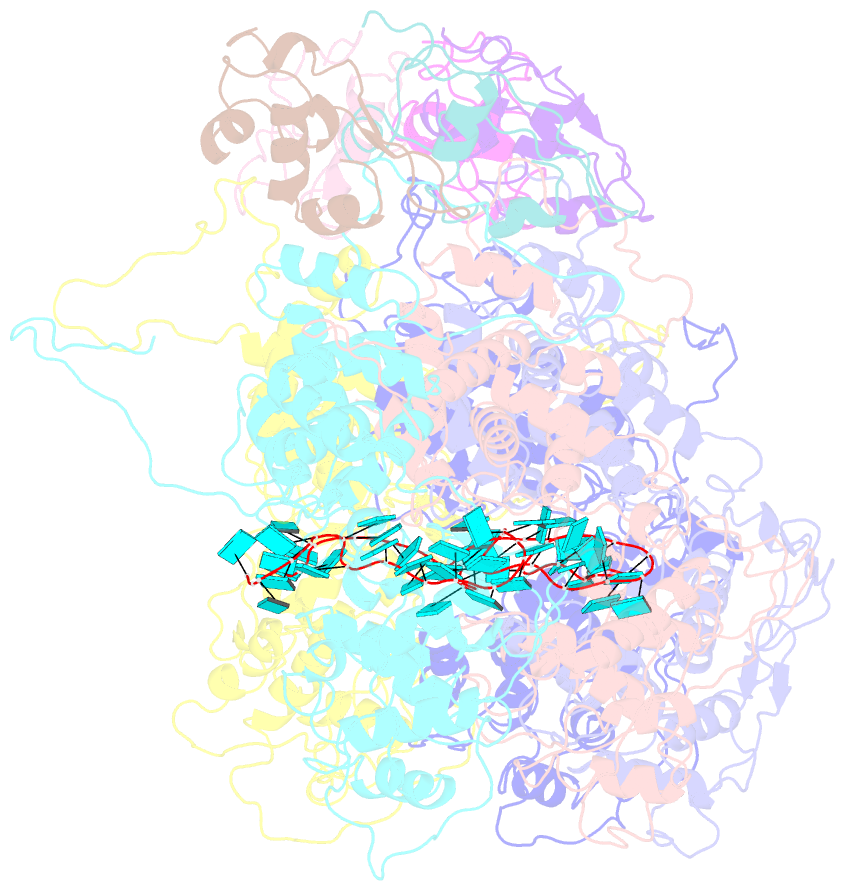
- Complex of the vesicular stomatitis virus nucleocapsid and the nucleocapsid-binding domain of the phosphoprotein
- Reference
- Green TJ, Luo M (2009): "Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P." Proc.Natl.Acad.Sci.USA, 106, 11713-11718. doi: 10.1073/pnas.0903228106.
- Abstract
- The negative-strand RNA viruses (NSRVs) are unique because their nucleocapsid, not the naked RNA, is the active template for transcription and replication. The viral polymerase of nonsegmented NSRVs contains a large polymerase catalytic subunit (L) and a nonenzymatic cofactor, the phosphoprotein (P). Insight into how P delivers the polymerase complex to the nucleocapsid has long been pursued by reverse genetics and biochemical approaches. Here, we present the X-ray crystal structure of the C-terminal domain of P of vesicular stomatitis virus, a prototypic nonsegmented NSRV, bound to nucleocapsid-like particles. P binds primarily to the C-terminal lobe of 2 adjacent N proteins within the nucleocapsid. This binding mode is exclusive to the nucleocapsid, not the nucleocapsid (N) protein in other existing forms. Localization of phosphorylation sites within P and their proximity to the RNA cavity give insight into how the L protein might be oriented to access the RNA template.