Summary information and primary citation
- PDB-id
- 3hou; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription,transferase-DNA-RNA hybrid
- Method
- X-ray (3.2 Å)
- Summary
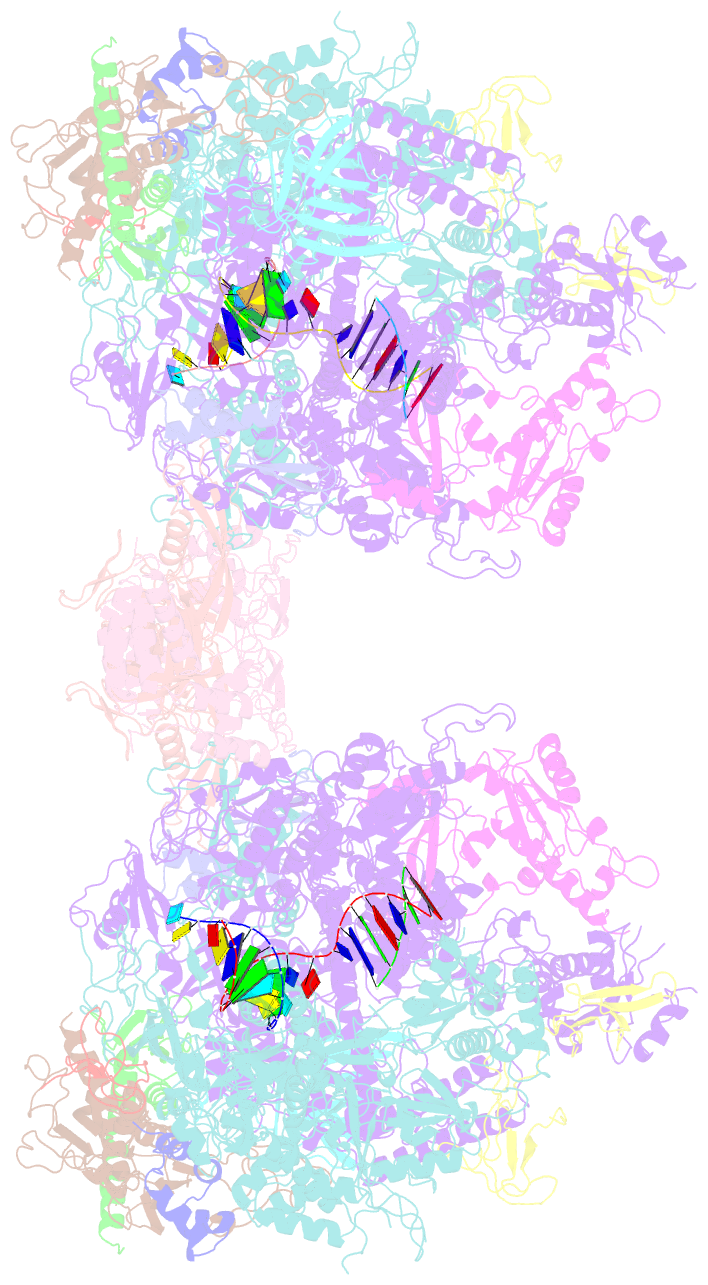
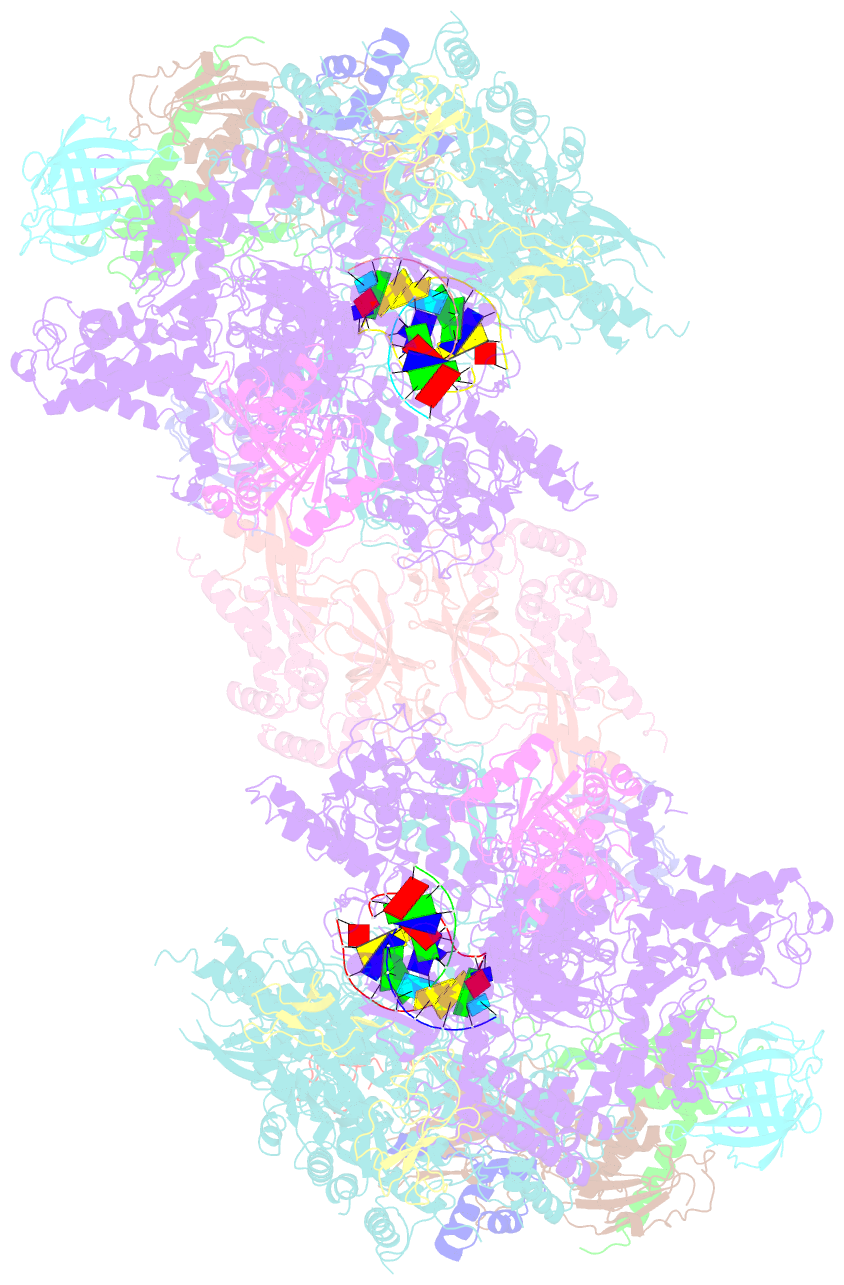
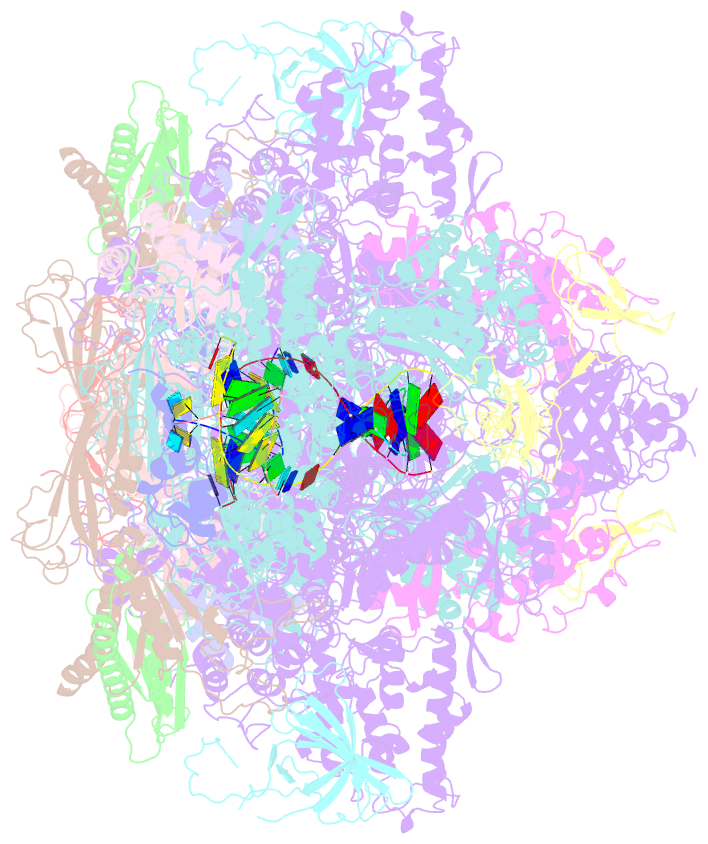
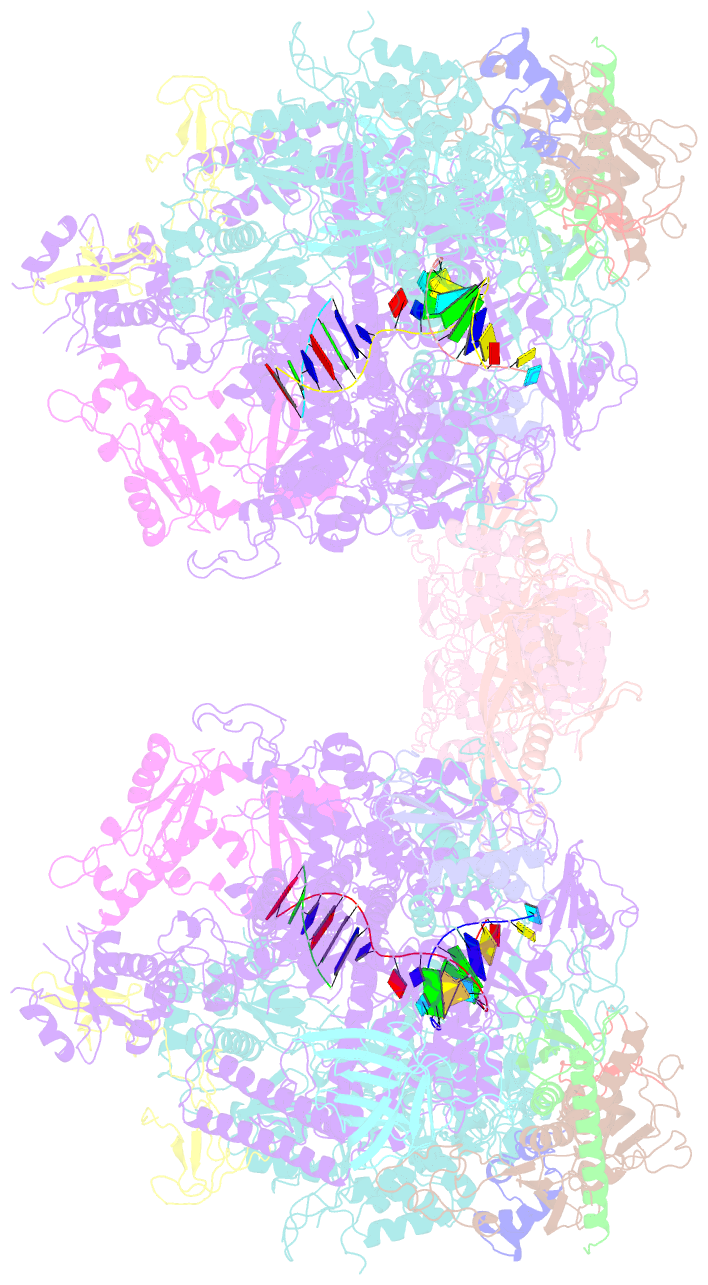
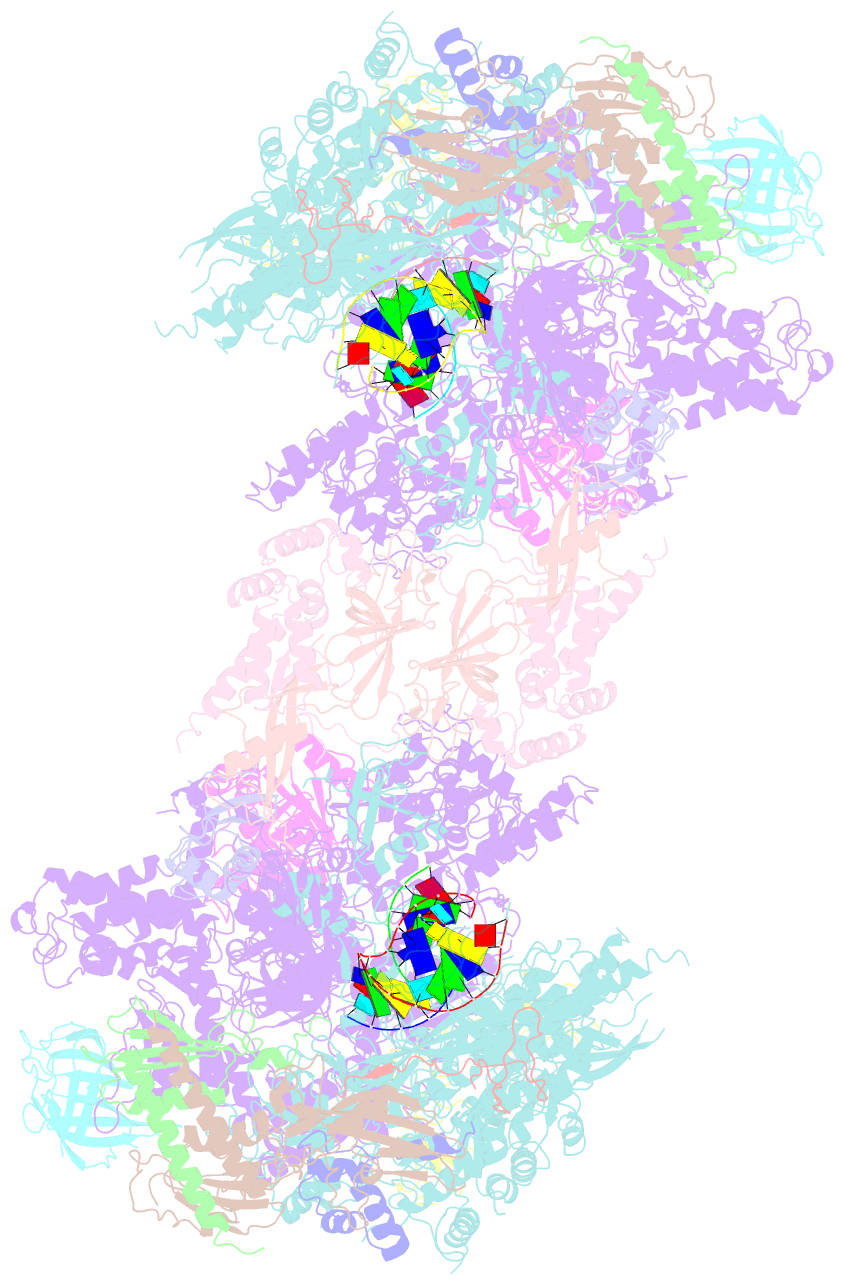
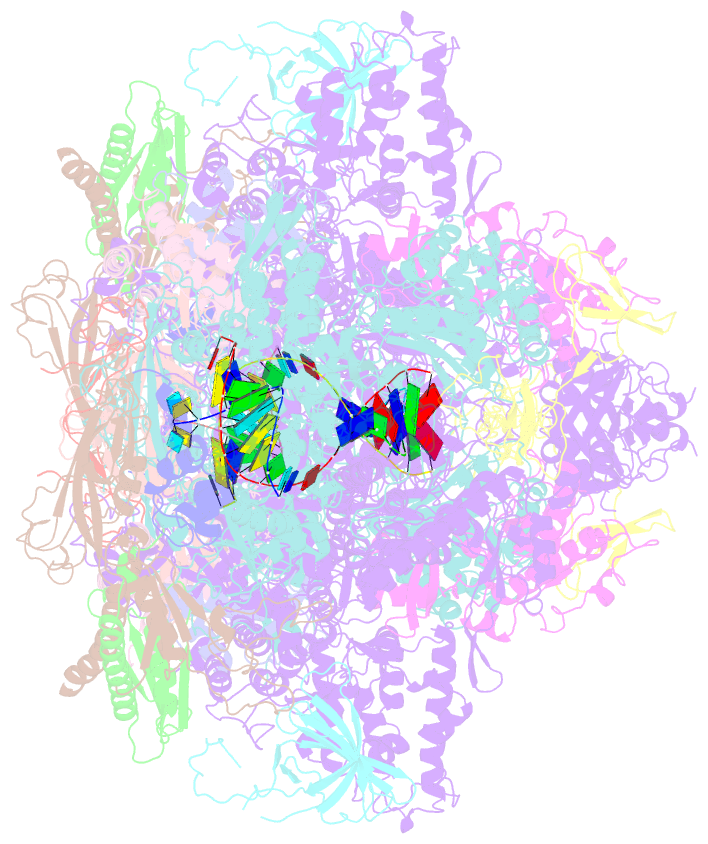
- Complete RNA polymerase ii elongation complex i with a t-u mismatch
- Reference
- Sydow JF, Brueckner F, Cheung ACM, Damsma GE, Dengl S, Lehmann E, Vassylyev D, Cramer P (2009): "Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA." Mol.Cell, 34, 710-721. doi: 10.1016/j.molcel.2009.06.002.
- Abstract
- We show that RNA polymerase (Pol) II prevents erroneous transcription in vitro with different strategies that depend on the type of DNARNA base mismatch. Certain mismatches are efficiently formed but impair RNA extension. Other mismatches allow for RNA extension but are inefficiently formed and efficiently proofread by RNA cleavage. X-ray analysis reveals that a TU mismatch impairs RNA extension by forming a wobble base pair at the Pol II active center that dissociates the catalytic metal ion and misaligns the RNA 3' end. The mismatch can also stabilize a paused state of Pol II with a frayed RNA 3' nucleotide. The frayed nucleotide binds in the Pol II pore either parallel or perpendicular to the DNA-RNA hybrid axis (fraying sites I and II, respectively) and overlaps the nucleoside triphosphate (NTP) site, explaining how it halts transcription during proofreading, before backtracking and RNA cleavage.