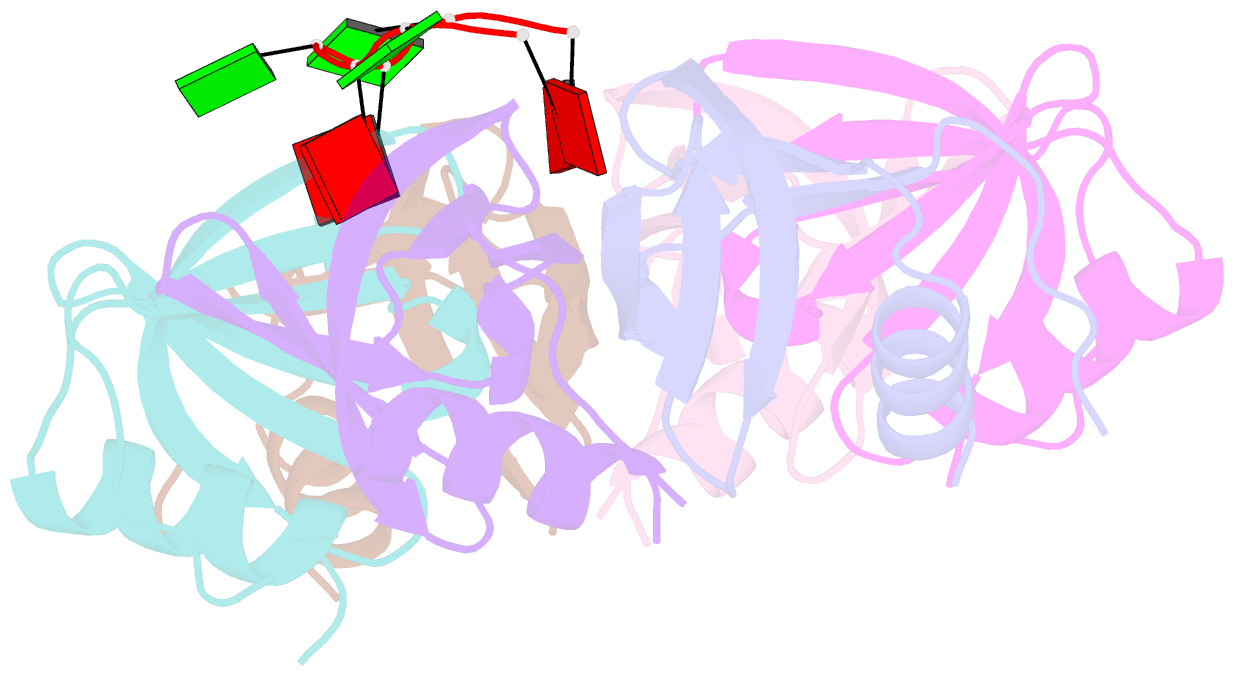
Summary information and primary citation
- PDB-id
- 3hsb; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (2.2 Å)
- Summary
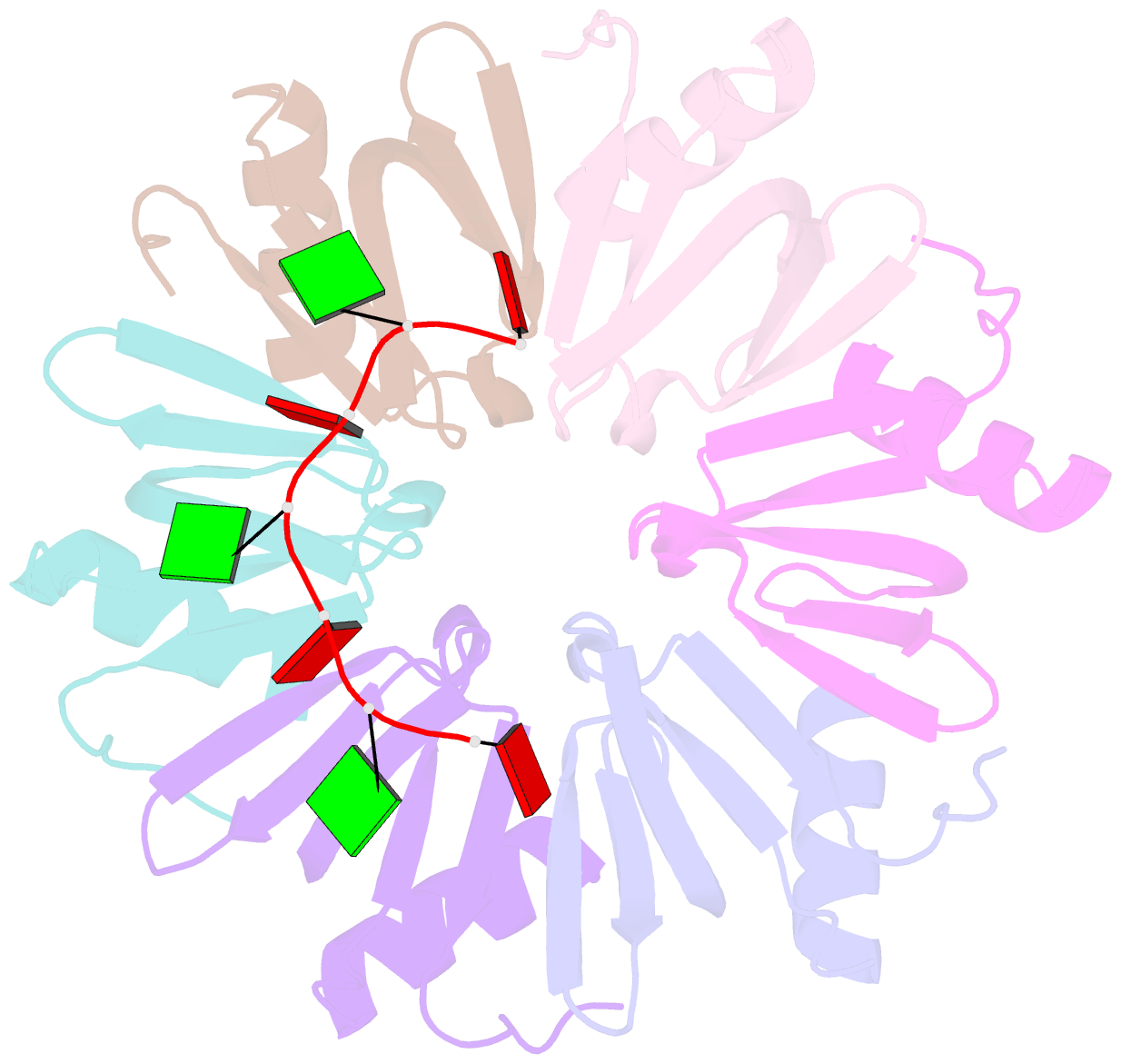
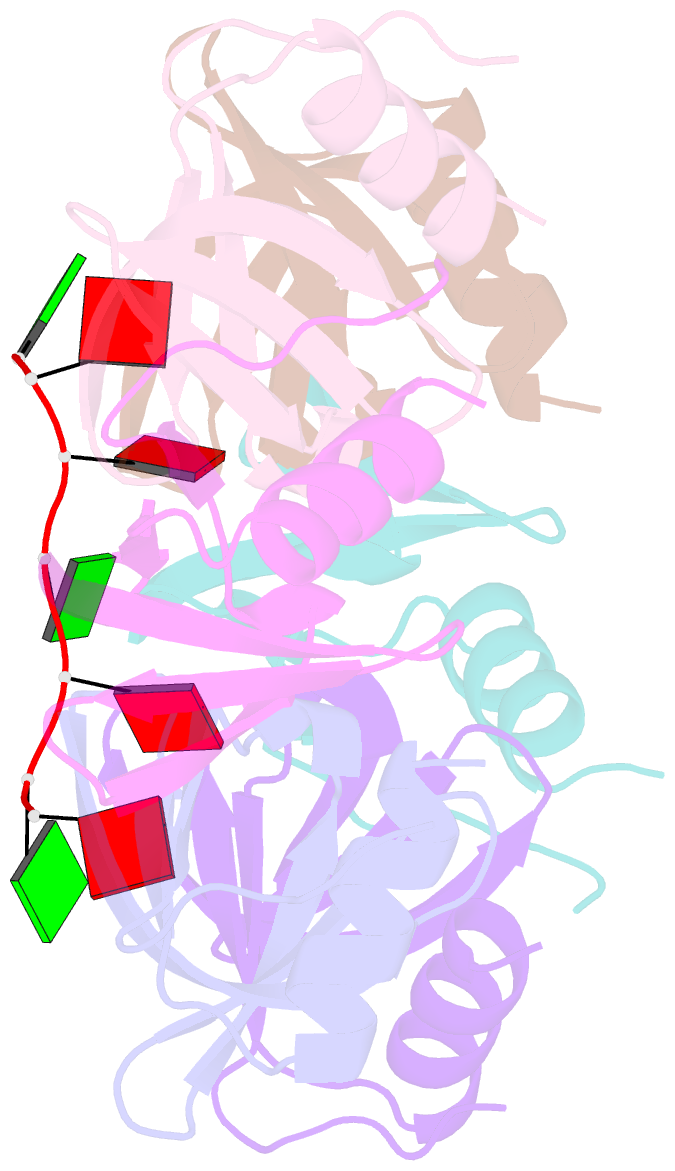
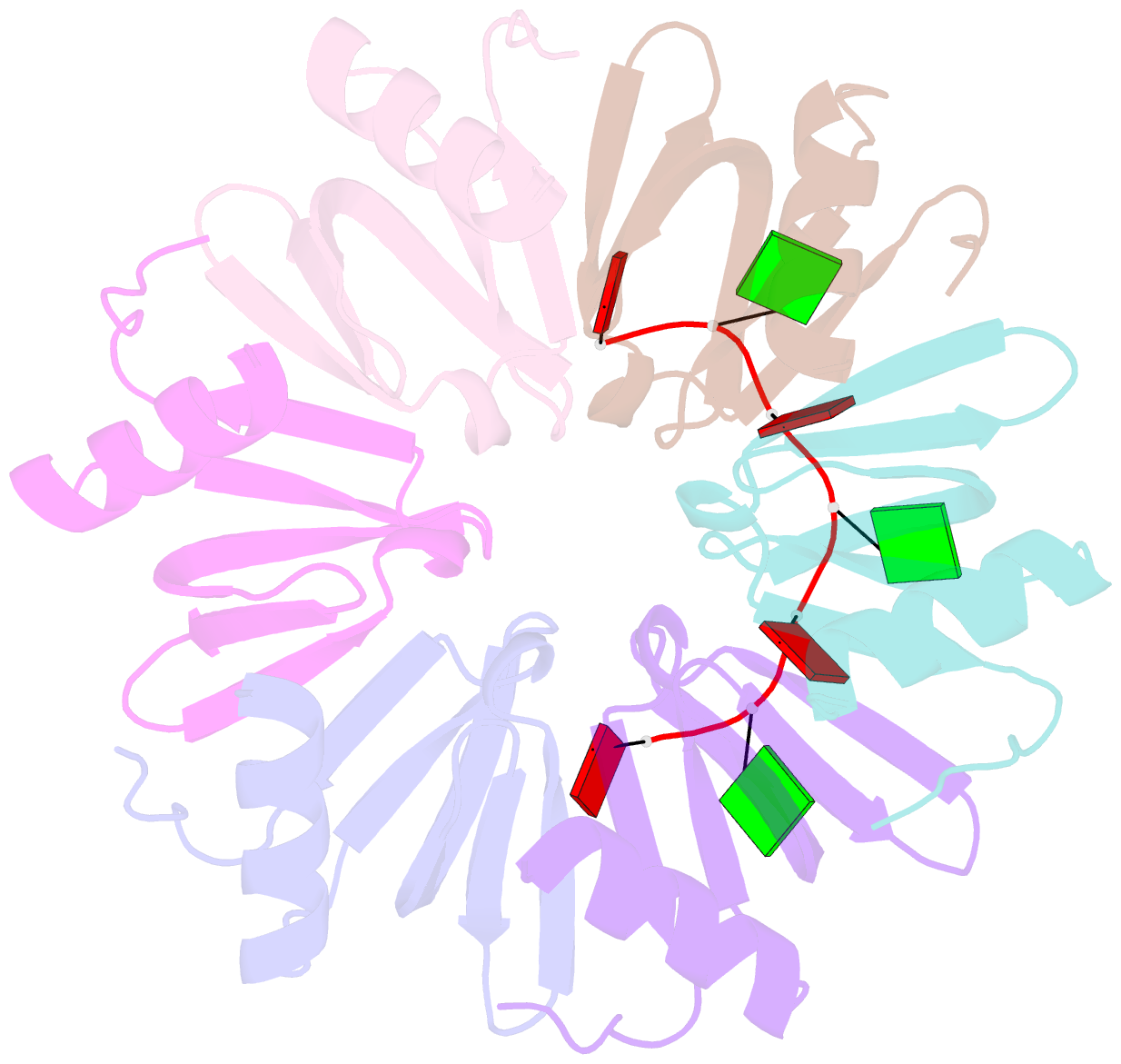
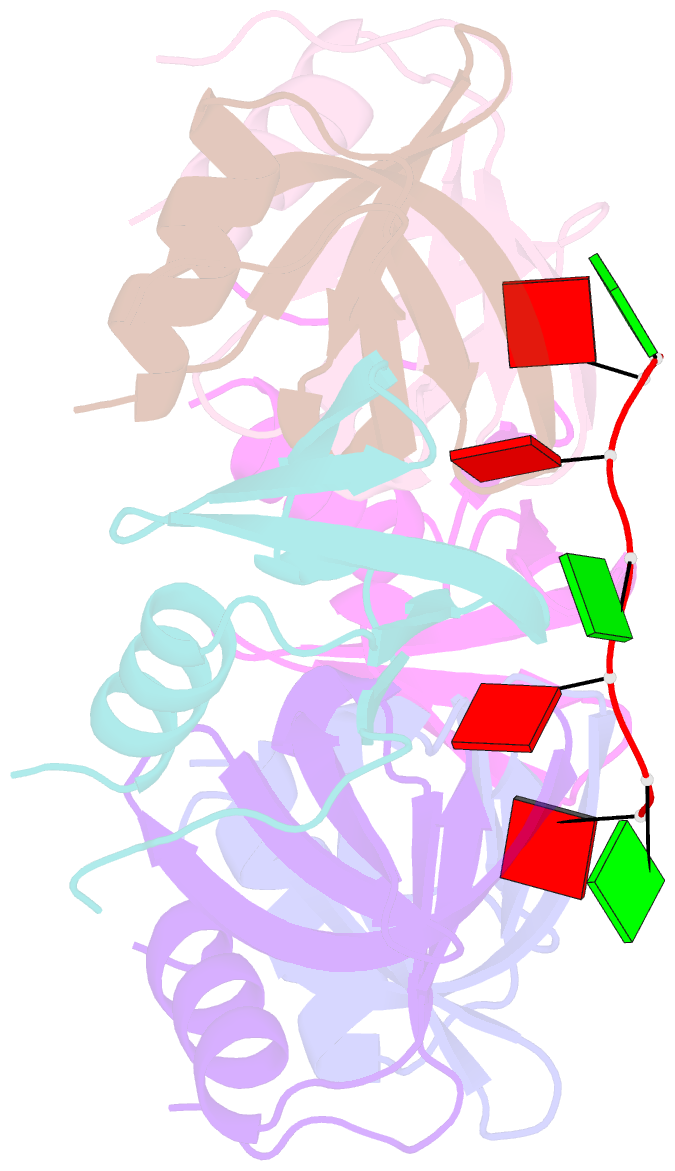
- Crystal structure of ymah (hfq) from bacillus subtilis in complex with an RNA aptamer
- Reference
- Someya T, Baba S, Fujimoto M, Kawai G, Kumasaka T, Nakamura K (2012): "Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: insight into RNA-binding properties of bacterial Hfq." Nucleic Acids Res., 40, 1856-1867. doi: 10.1093/nar/gkr892.
- Abstract
- Bacterial Hfq is a protein that plays an important role in the regulation of genes in cooperation with sRNAs. Escherichia coli Hfq (EcHfq) has two or more sites that bind RNA(s) including U-rich and/or the poly(A) tail of mRNA. However, functional and structural information about Bacillus subtilis Hfq (BsHfq) including the RNA sequences that specifically bind to it remain unknown. Here, we describe RNA aptamers including fragment (AG)(3)A that are recognized by BsHfq and crystal structures of the BsHfq-(AG)(3)A complex at 2.2 Å resolution. Mutational and structural studies revealed that the RNA fragment binds to the distal site, one of the two binding sites on Hfq, and identified amino acid residues that are critical for sequence-specific interactions between BsHfq and (AG)(3)A. In particular, R32 appears to interact with G bases in (AG)(3)A. Poly(A) also binds to the distal site of EcHfq, but the overall RNA structure and protein-RNA interaction patterns engaged in the R32 residues of BsHfq-(AG)(3)A differ from those of EcHfq-poly(A). These findings provide novel insight into how the Hfq homologue recognizes RNA.