Summary information and primary citation
- PDB-id
- 3i0x; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase,lyase-DNA
- Method
- X-ray (1.8 Å)
- Summary
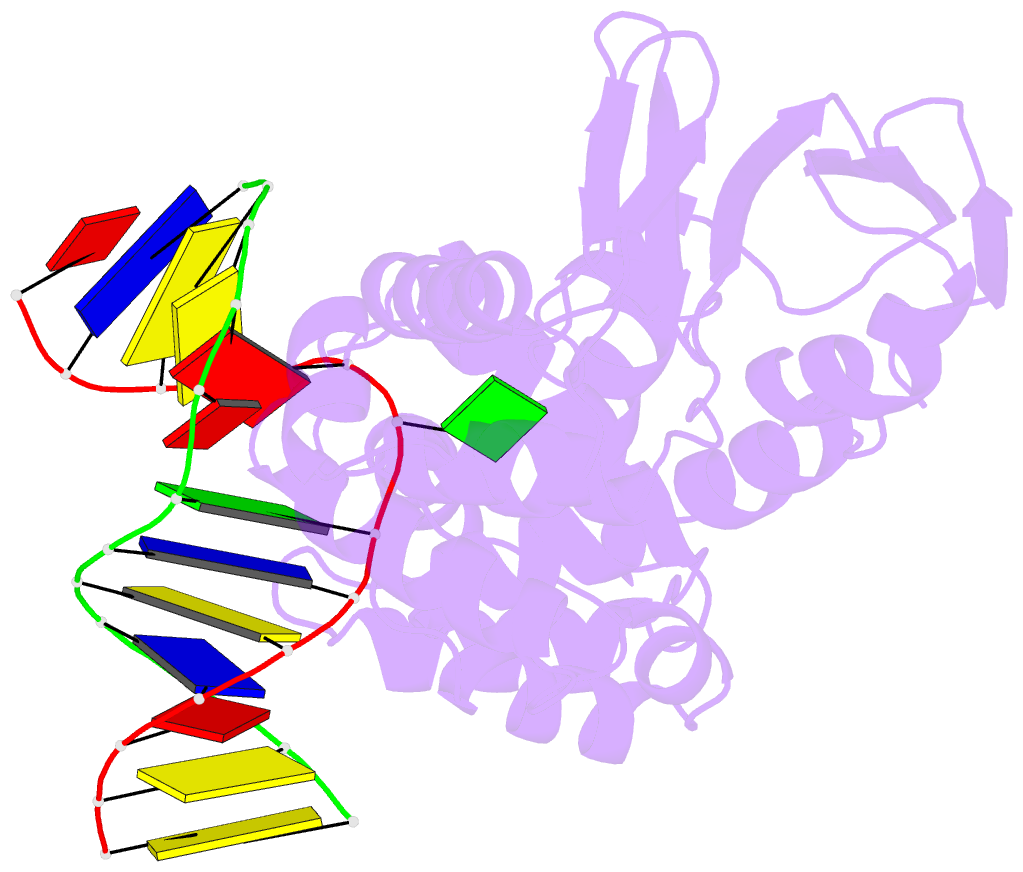
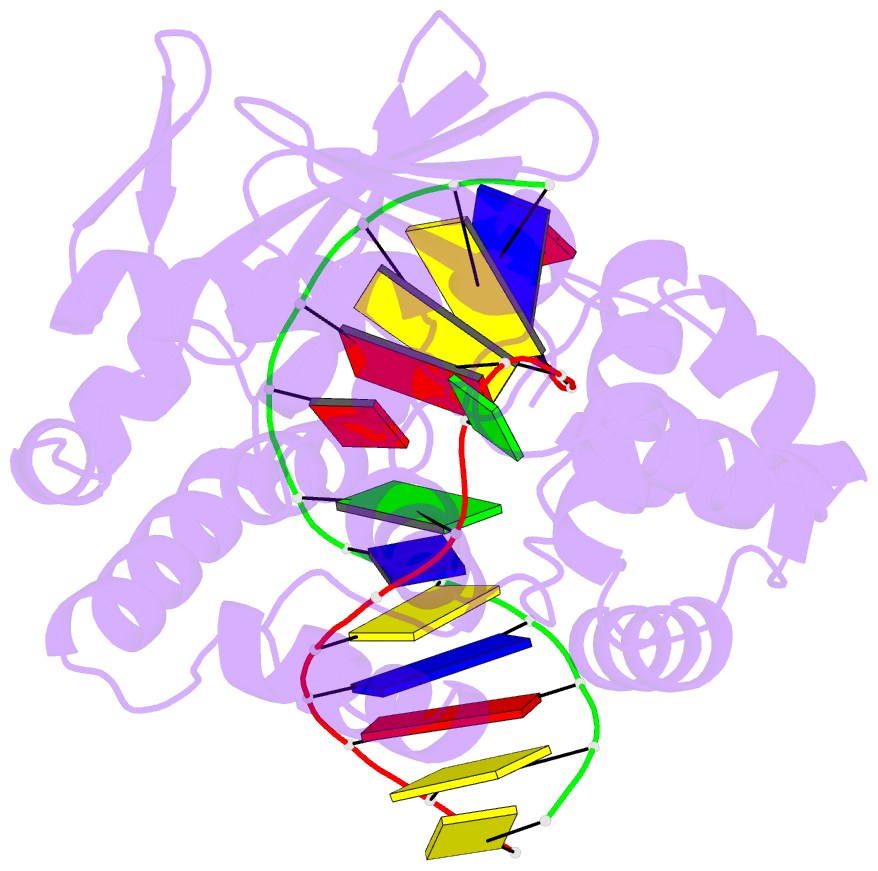
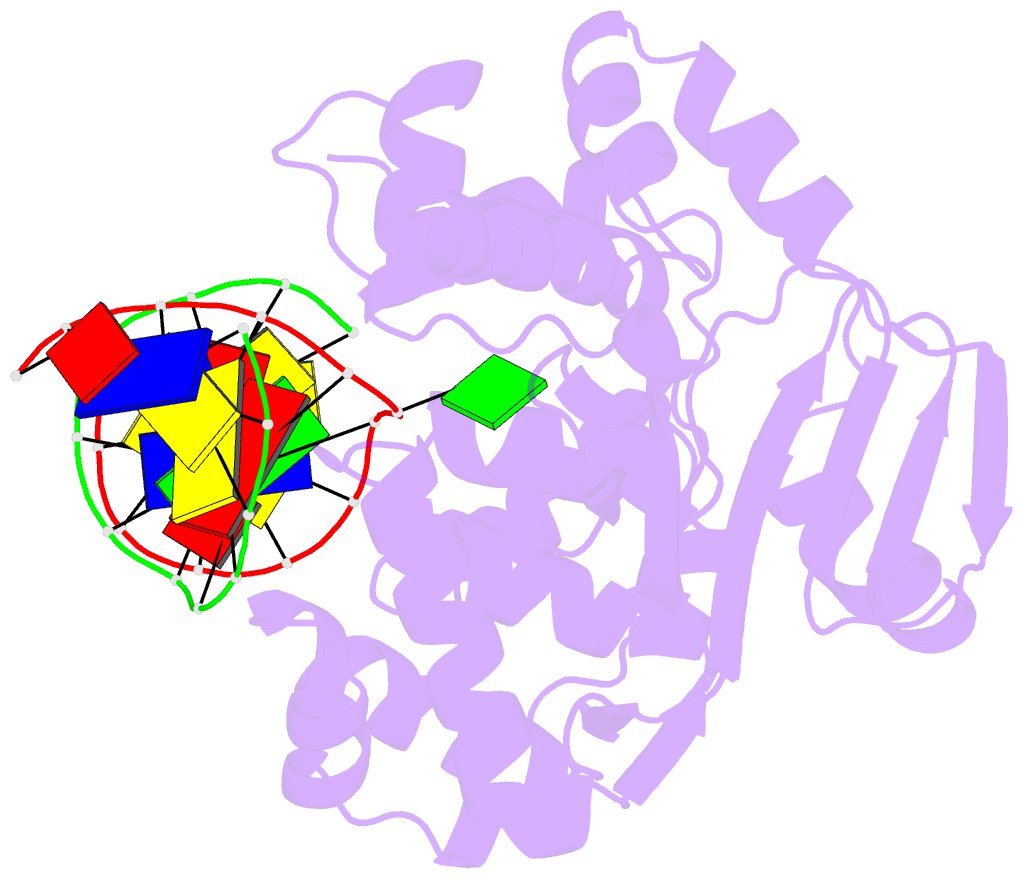
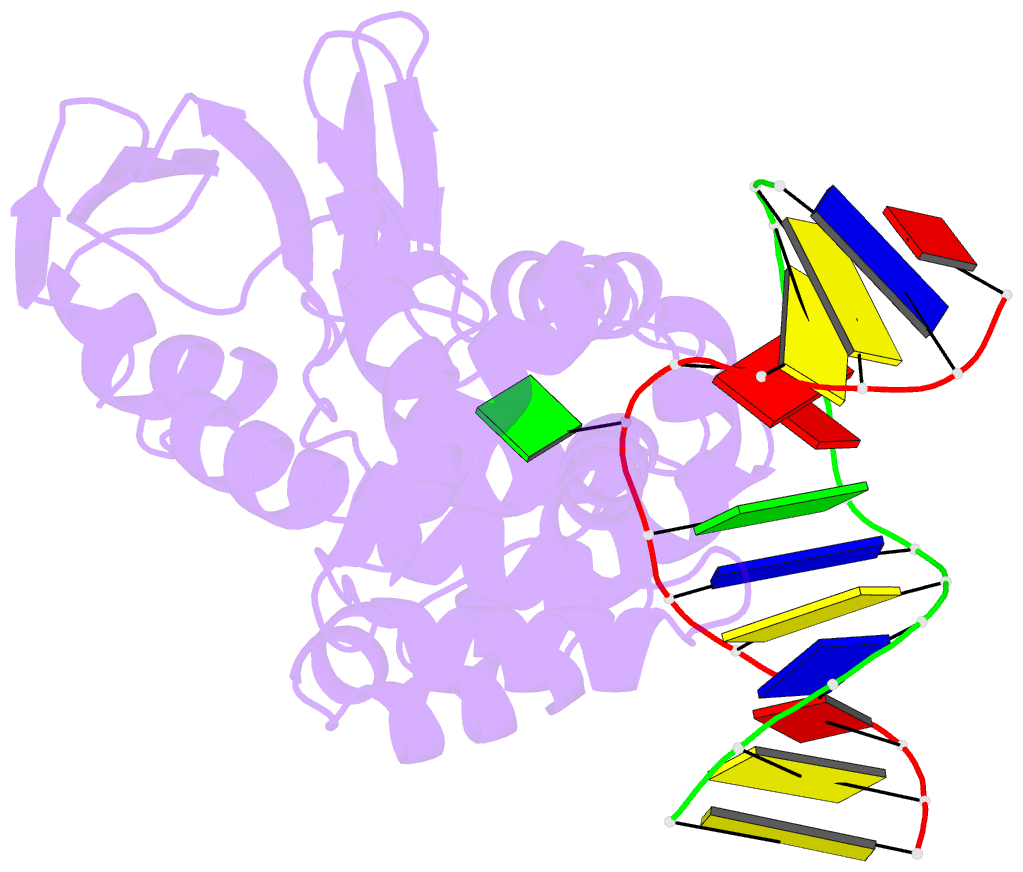
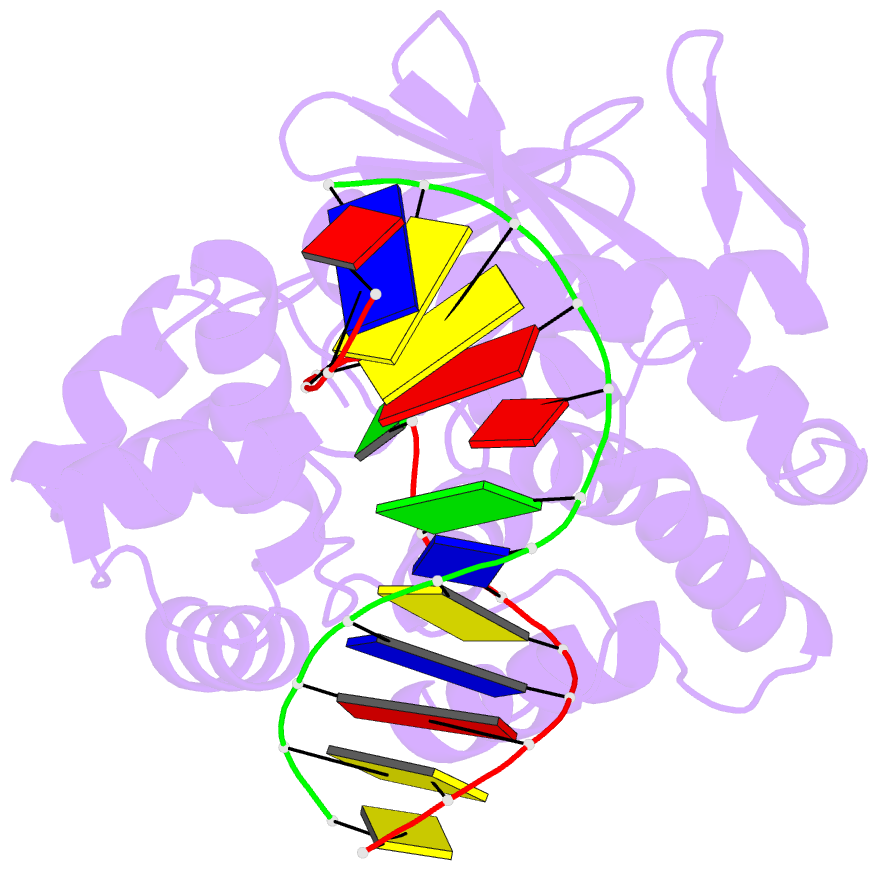
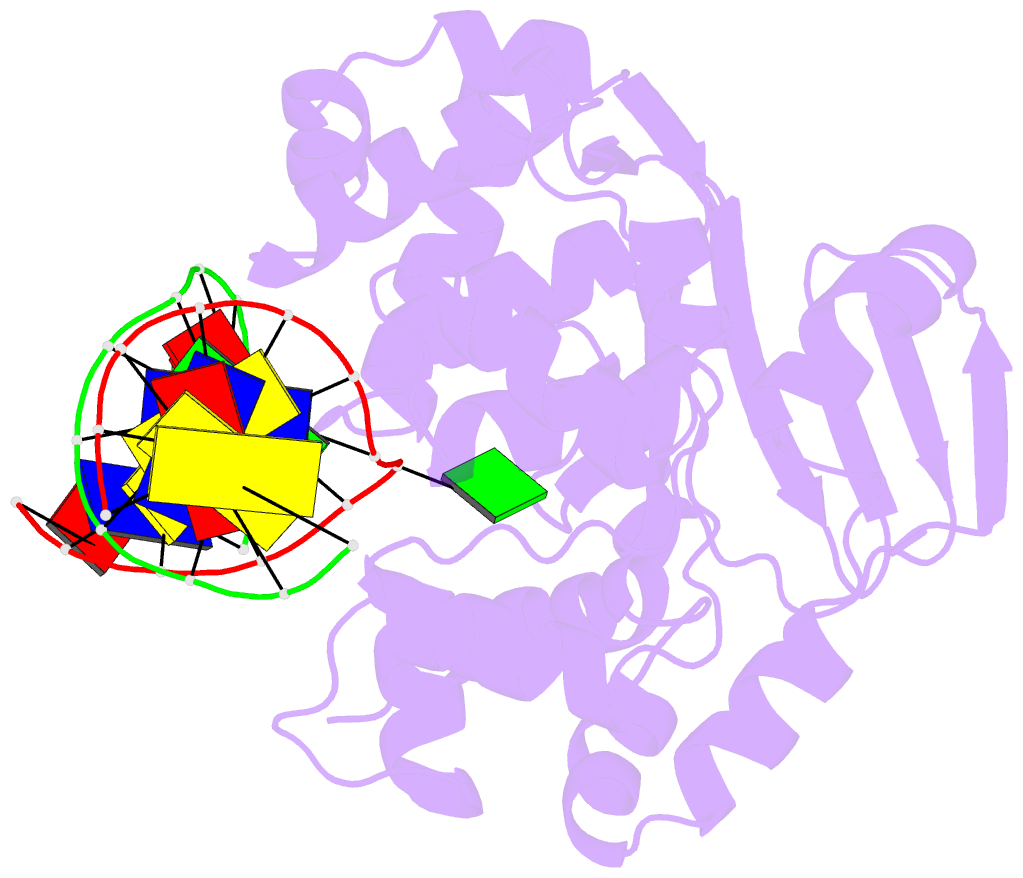
- Crystal structure of clostridium acetobutylicum 8-oxoguanine glycosylase-lyase in complex with dsDNA containing adenine opposite to 8-oxog
- Reference
- Faucher F, Wallace SS, Doublie S (2009): "Structural basis for the lack of opposite base specificity of Clostridium acetobutylicum 8-oxoguanine DNA glycosylase." Dna Repair, 8, 1283-1289. doi: 10.1016/j.dnarep.2009.08.002.
- Abstract
- 7,8-Dihydro-8-oxoguanine (8-oxoG) is the major oxidative product of guanine and the most prevalent base lesion observed in DNA molecules. Because 8-oxoG has the capability to form a Hoogsteen pair with adenine (8-oxoG:A) in addition to a normal Watson-Crick pair with cytosine (8-oxoG:C), this lesion can lead to a G:C-->T:A transversion after replication. However, 8-oxoG is recognized and excised by the 8-oxoguanine DNA glycosylase (Ogg) of the base excision repair pathway. Members of the Ogg1 family usually display a strong preference for a C opposite the lesion. In contrast, the atypical Ogg1 from Clostridium actetobutylicum (CacOgg) can excise 8-oxoG when paired with either one of the four bases, albeit with a preference for C and A. Here we describe the first high-resolution crystal structures of CacOgg in complex with duplex DNA containing the 8-oxoG lesion paired to cytosine and to adenine. A structural comparison with human OGG1 provides a rationale for the lack of opposite base specificity displayed by the bacterial Ogg.