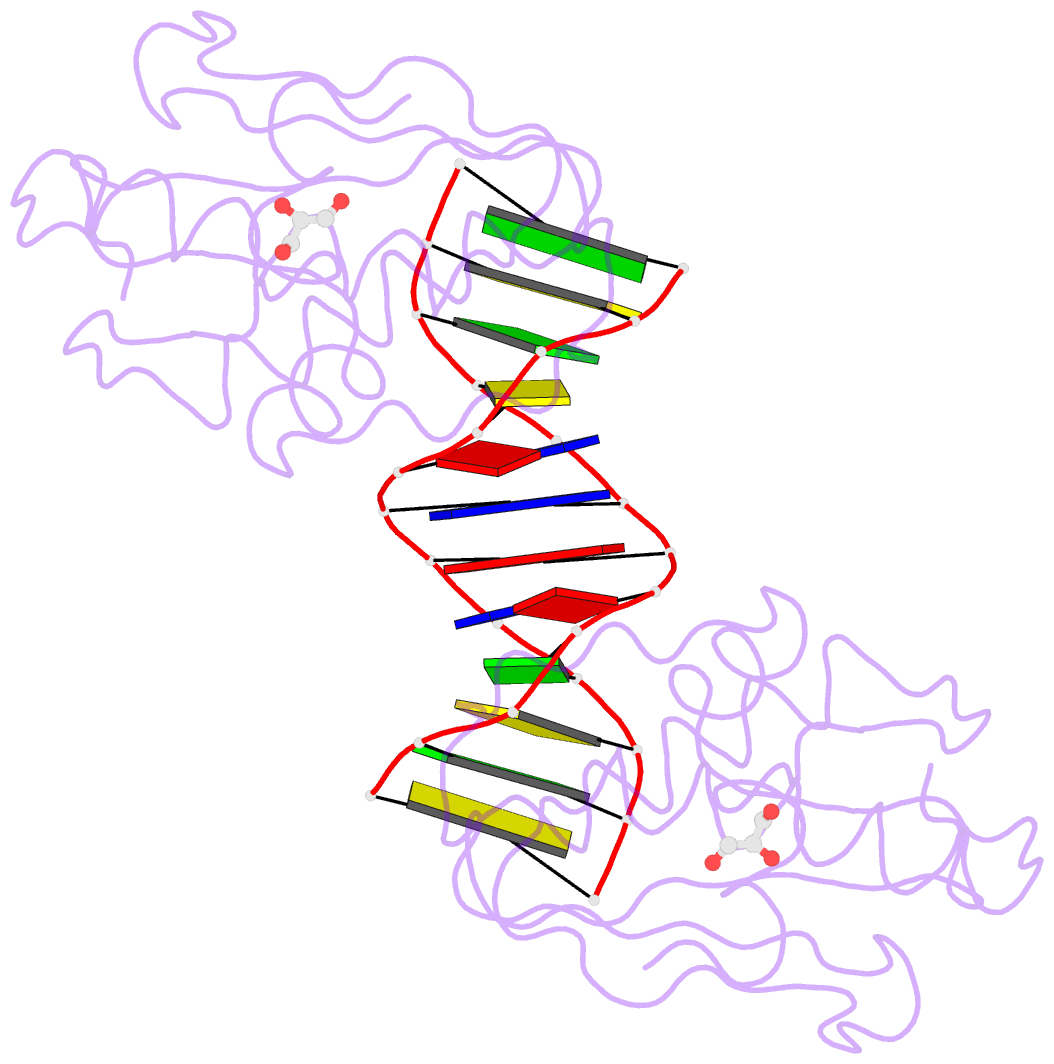
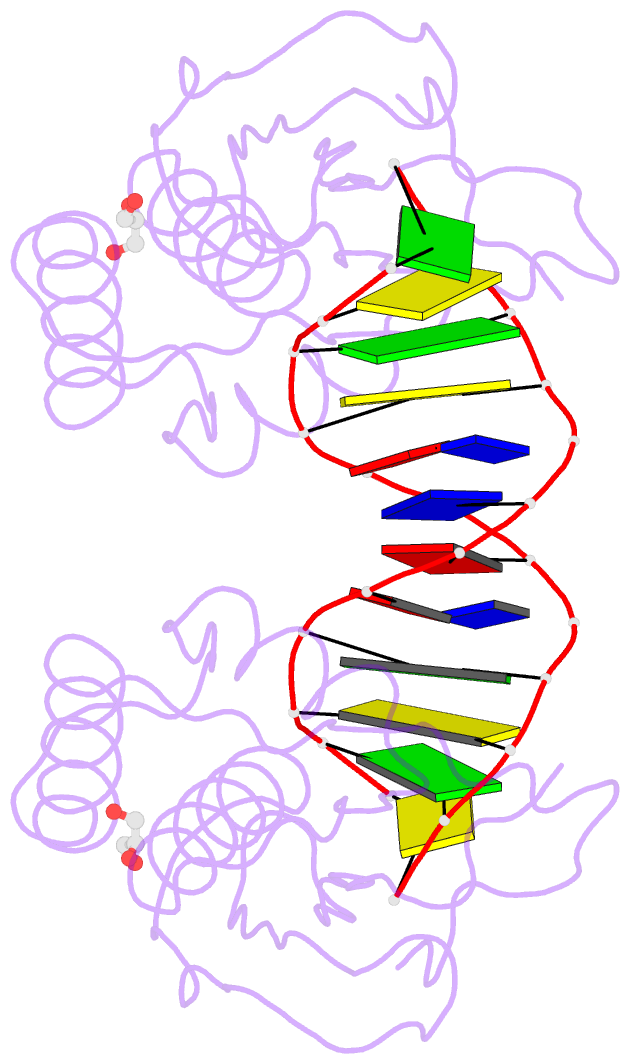
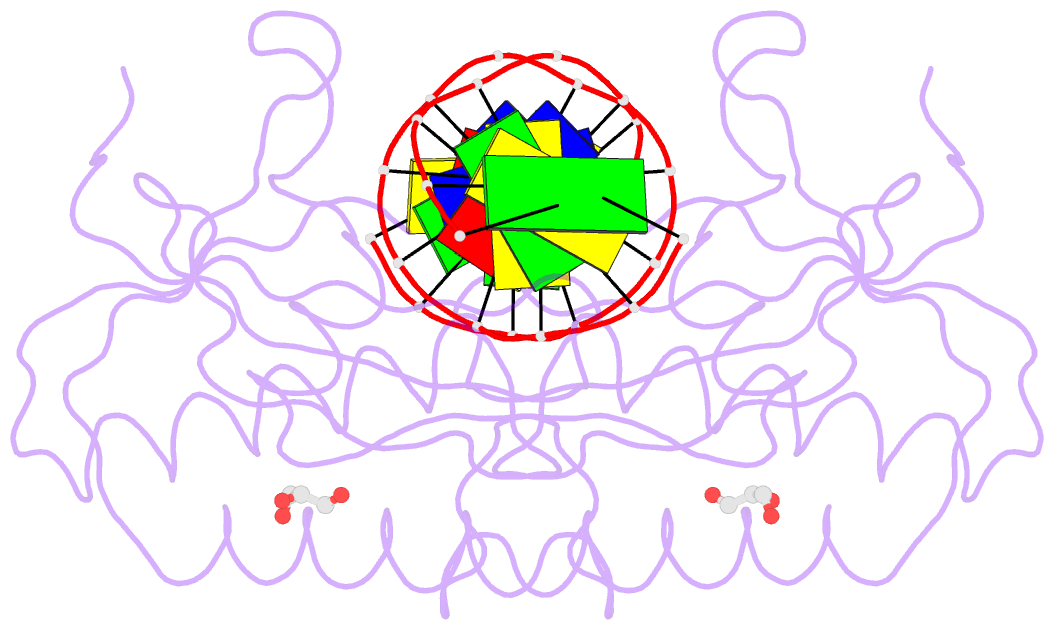
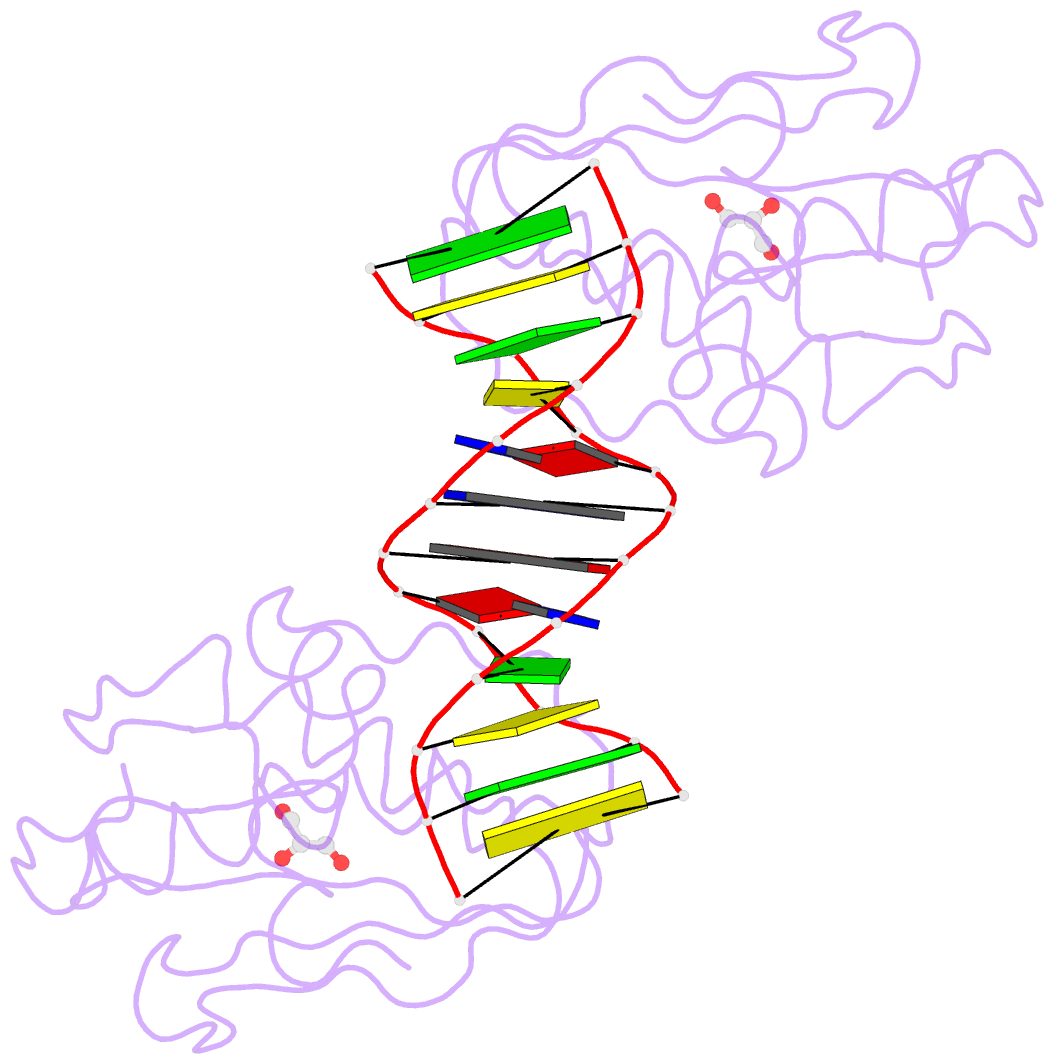
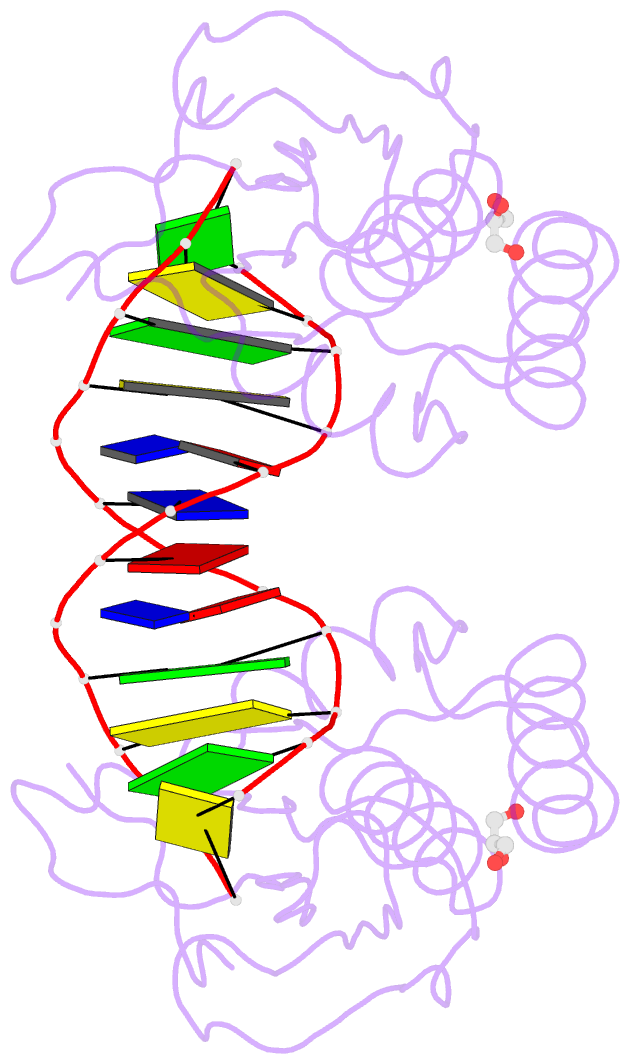
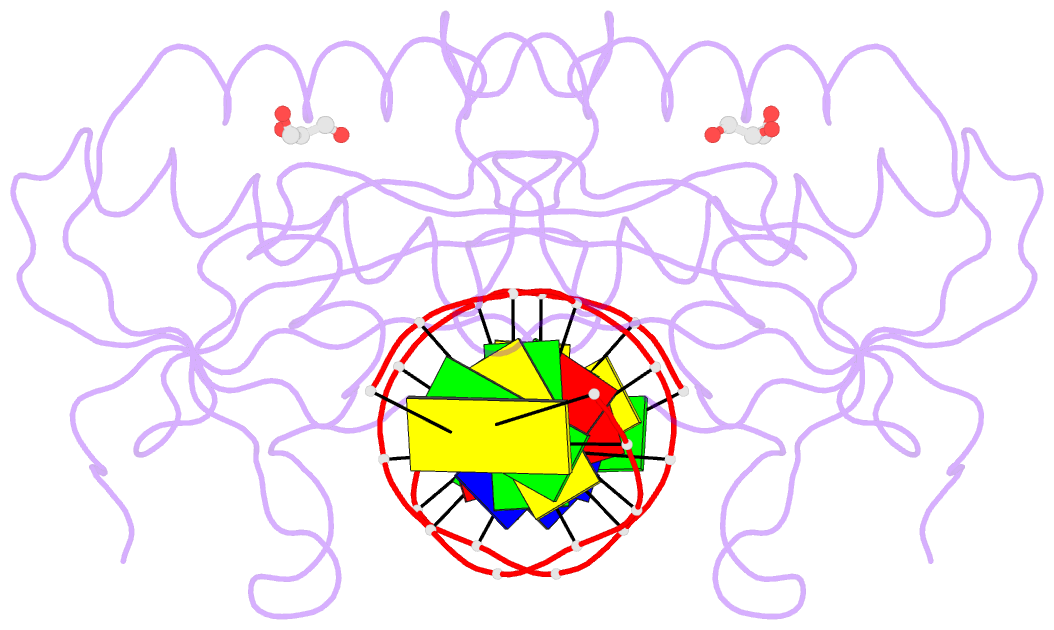
Summary information and primary citation
- PDB-id
- 3i8d; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.61 Å)
- Summary
- The pairing geometry of the hydrophobic thymine analog 2,4-difluorotoluene in duplex DNA as analyzed by x-ray crystallography
- Reference
- Pallan PS, Egli M (2009): "Pairing geometry of the hydrophobic thymine analogue 2,4-difluorotoluene in duplex DNA as analyzed by X-ray crystallography." J.Am.Chem.Soc., 131, 12548-12549. doi: 10.1021/ja905739j.
- Abstract
- Certain DNA polymerases (pols) were found to efficiently insert A opposite the hydrophobic T isostere 2,4-difluorotoluene (F) and vice versa, resulting in the widely held belief that some pols rely on shape rather than H-bonding for accurate replication. Using X-ray crystallography we have analyzed the geometry of F:A pairs in duplex DNA and observed a distance between fluorine and the exocyclic amino group of A that is consistent with a H-bond, thus challenging the assumption that the F analogue is unable to engage in H-bonding as well as the steric hypothesis of DNA replication. Therefore, shape and H-bonding are inherently related, and steric constraints at a pol active site, or conferred by stacking or the DNA backbone conformation, may enable H-bonding by F.