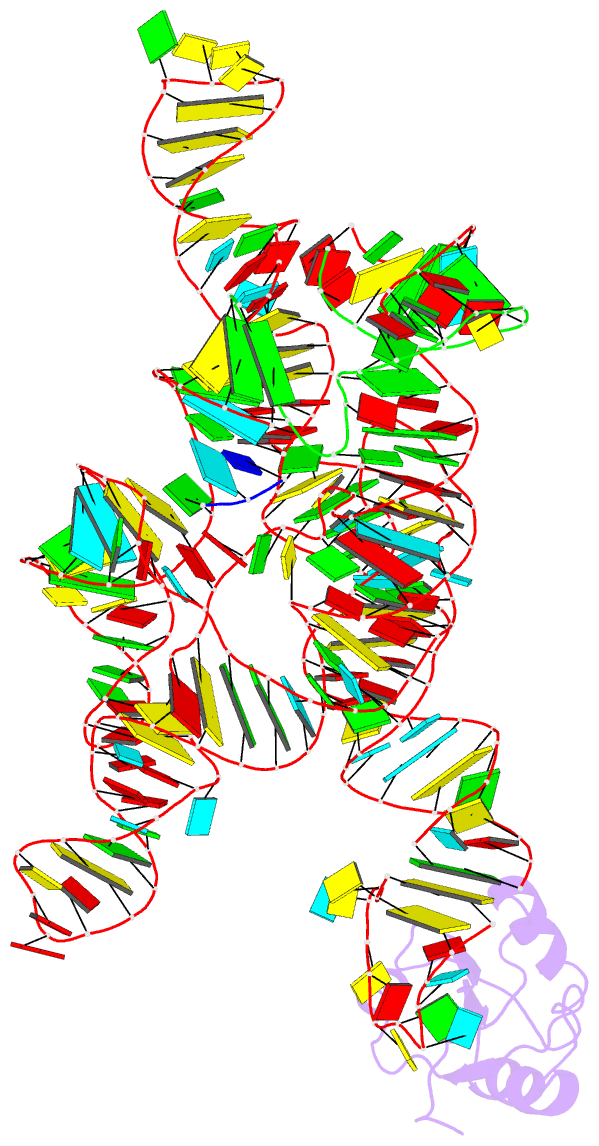
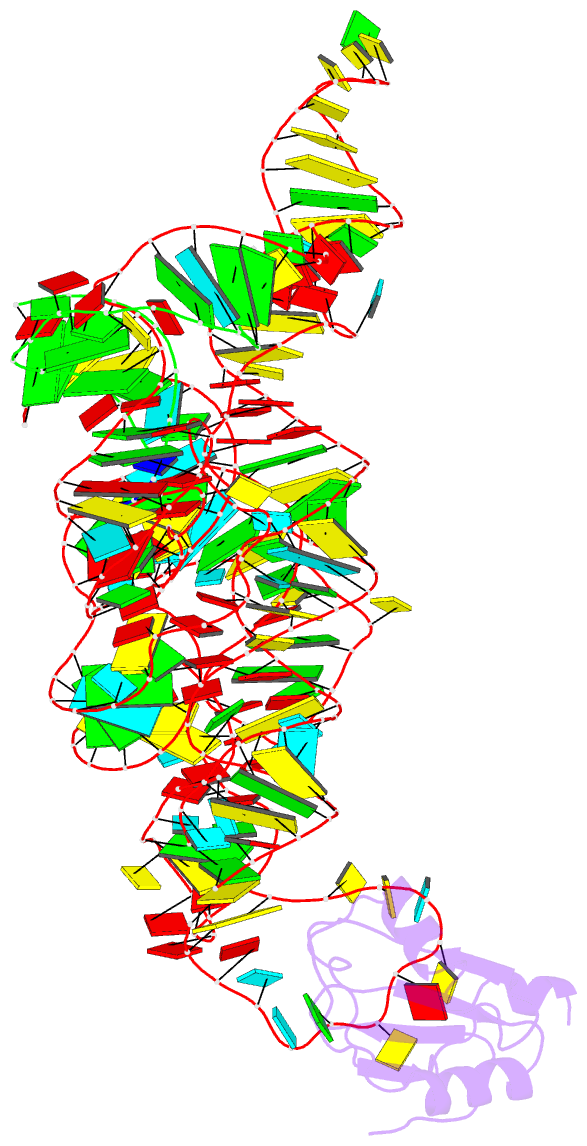
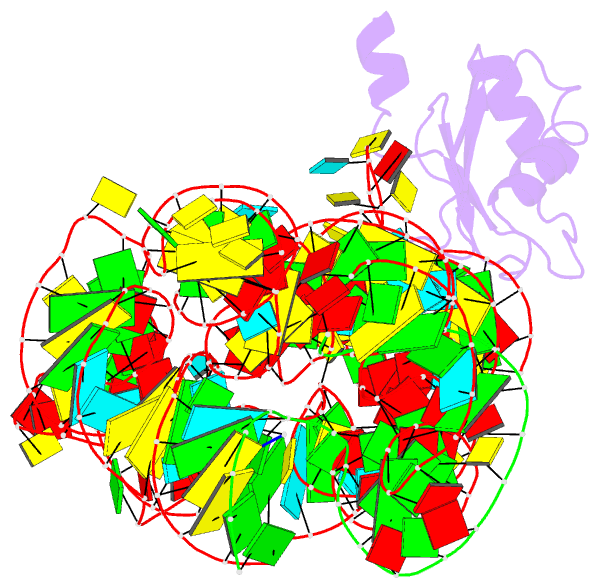
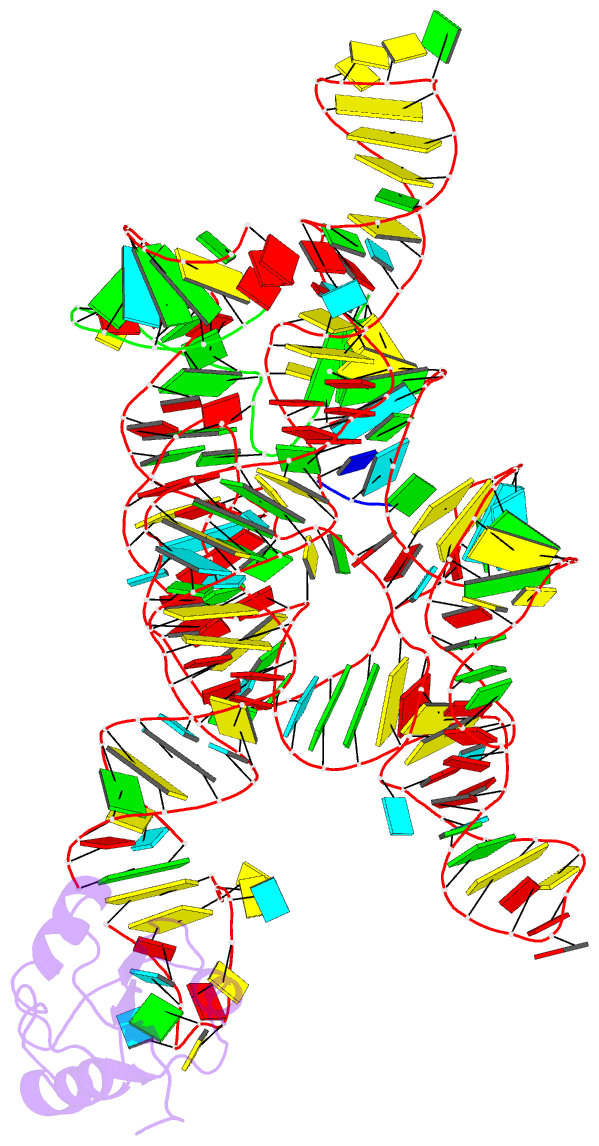
Summary information and primary citation
- PDB-id
- 3iin; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-DNA-RNA
- Method
- X-ray (4.18 Å)
- Summary
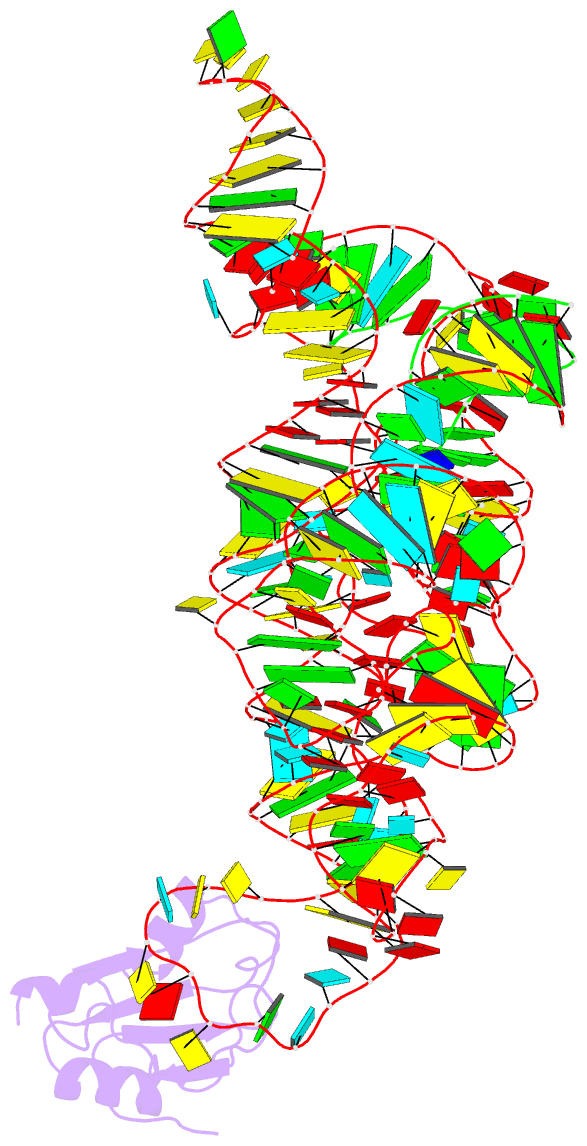
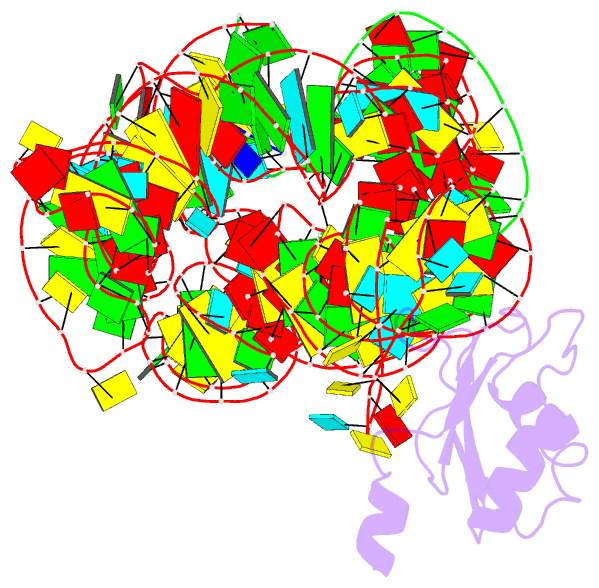
- Plasticity of the kink turn structural motif
- Reference
- Antonioli AH, Cochrane JC, Lipchock SV, Strobel SA (2010): "Plasticity of the RNA kink turn structural motif." Rna, 16, 762-768. doi: 10.1261/rna.1883810.
- Abstract
- The kink turn (K-turn) is an RNA structural motif found in many biologically significant RNAs. While most examples of the K-turn have a similar fold, the crystal structure of the Azoarcus group I intron revealed a novel RNA conformation, a reverse kink turn bent in the direction opposite that of a consensus K-turn. The reverse K-turn is bent toward the major grooves rather than the minor grooves of the flanking helices, yet the sequence differs from the K-turn consensus by only a single nucleotide. Here we demonstrate that the reverse bend direction is not solely defined by internal sequence elements, but is instead affected by structural elements external to the K-turn. It bends toward the major groove under the direction of a tetraloop-tetraloop receptor. The ability of one sequence to form two distinct structures demonstrates the inherent plasticity of the K-turn sequence. Such plasticity suggests that the K-turn is not a primary element in RNA folding, but instead is shaped by other structural elements within the RNA or ribonucleoprotein assembly.