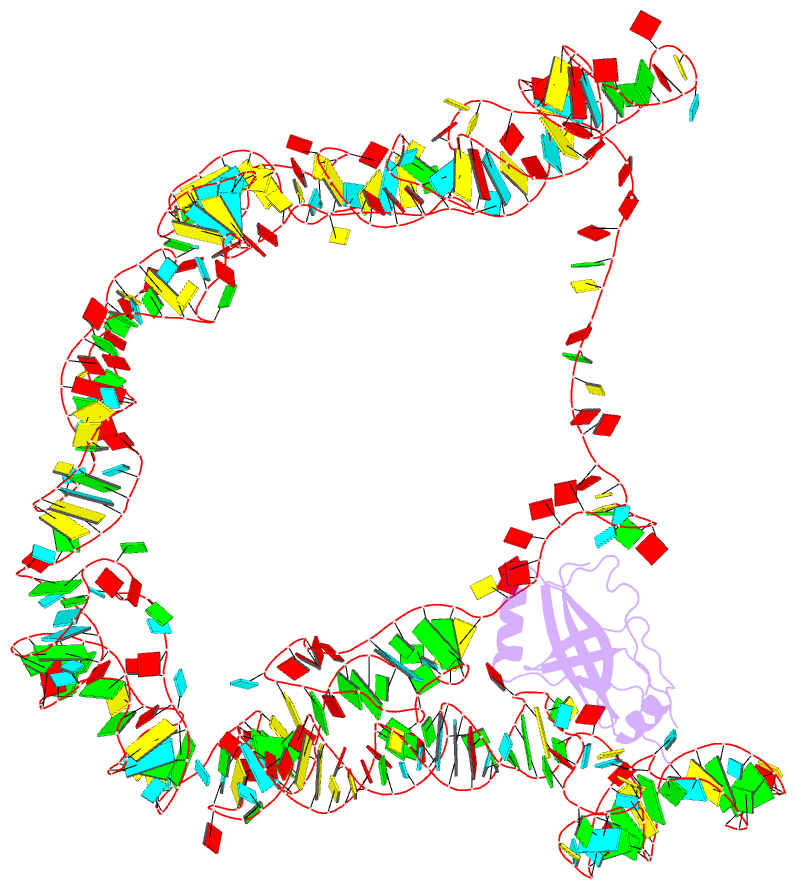
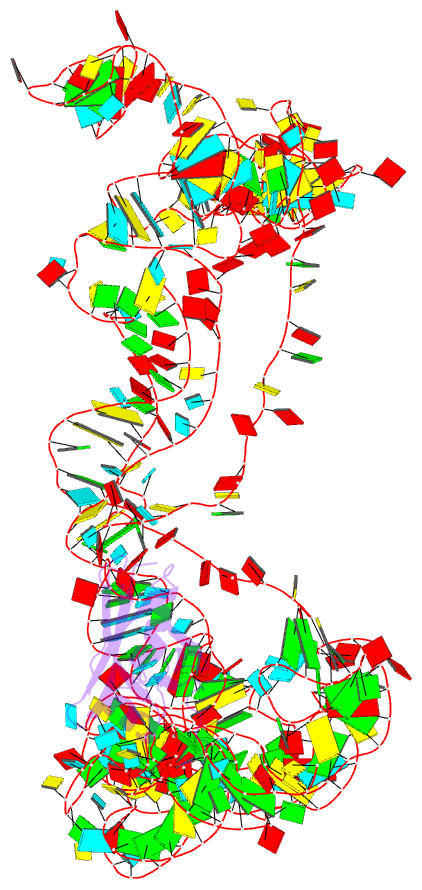
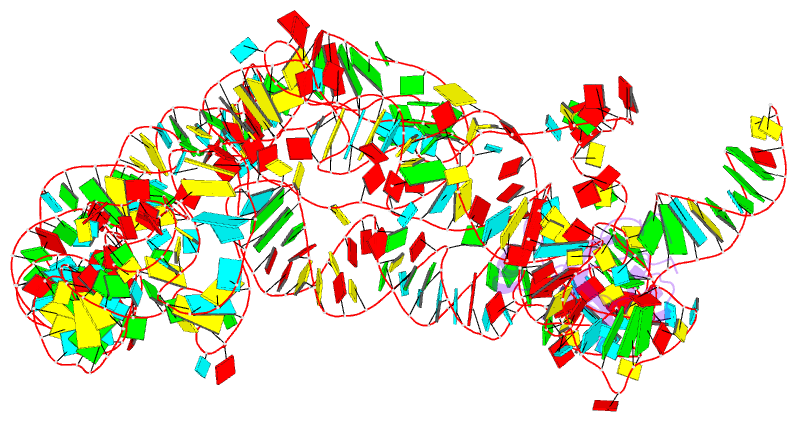
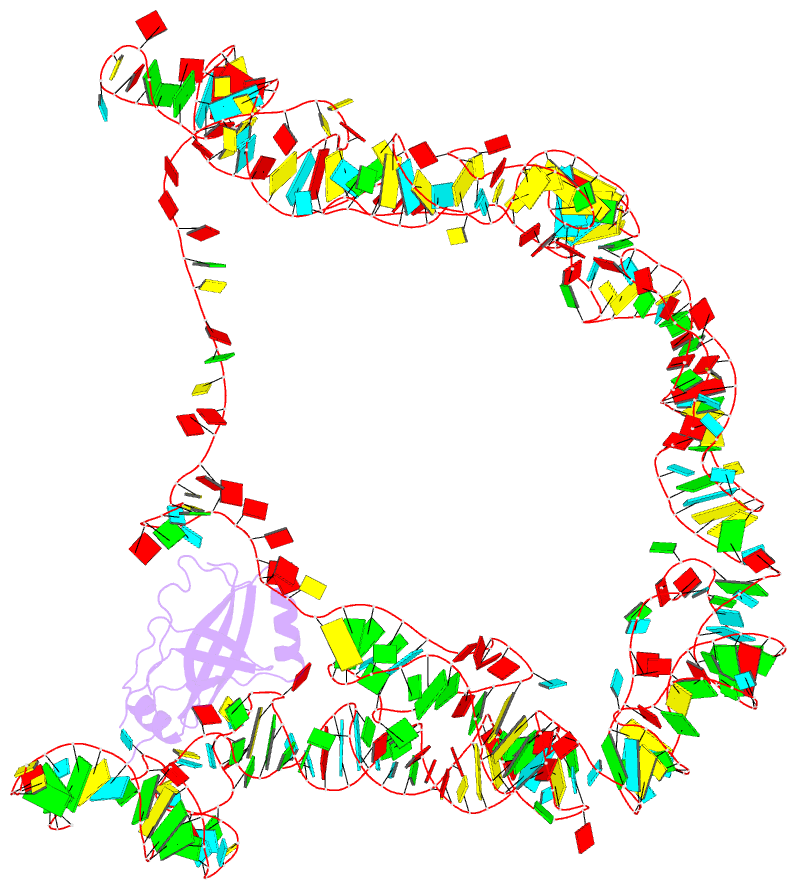
Summary information and primary citation
- PDB-id
- 3iz4; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- cryo-EM (13.6 Å)
- Summary
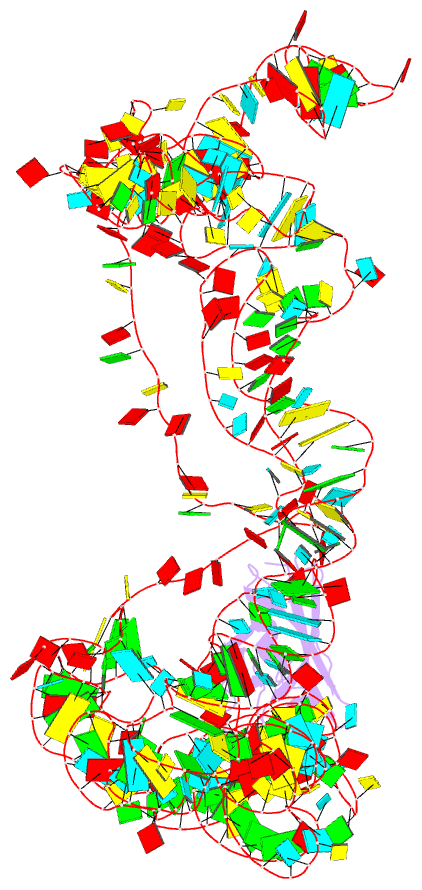
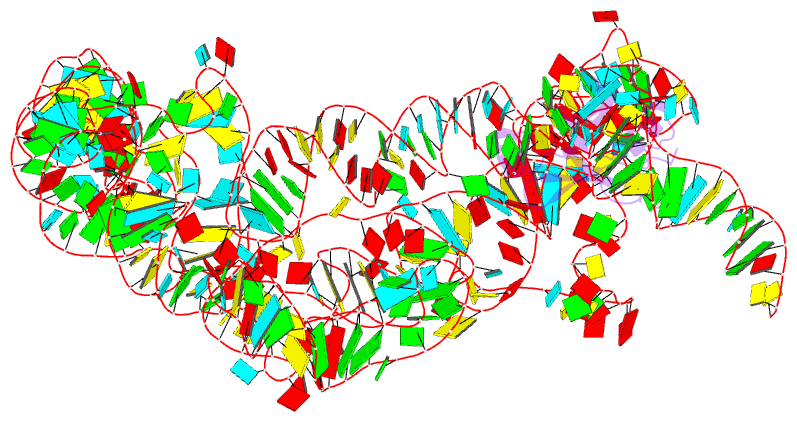
- Modified e. coli tmrna in the resume state with the trna-like domain in the ribosomal p site interacting with the smpb
- Reference
- Fu J, Hashem Y, Wower I, Lei J, Liao HY, Zwieb C, Wower J, Frank J (2010): "Visualizing the transfer-messenger RNA as the ribosome resumes translation." Embo J., 29, 3819-3825. doi: 10.1038/emboj.2010.255.
- Abstract
- Bacterial ribosomes stalled by truncated mRNAs are rescued by transfer-messenger RNA (tmRNA), a dual-function molecule that contains a tRNA-like domain (TLD) and an internal open reading frame (ORF). Occupying the empty A site with its TLD, the tmRNA enters the ribosome with the help of elongation factor Tu and a protein factor called small protein B (SmpB), and switches the translation to its own ORF. In this study, using cryo-electron microscopy, we obtained the first structure of an in vivo-formed complex containing ribosome and the tmRNA at the point where the TLD is accommodated into the ribosomal P site. We show that tmRNA maintains a stable 'arc and fork' structure on the ribosome when its TLD moves to the ribosomal P site and translation resumes on its ORF. Based on the density map, we built an atomic model, which suggests that SmpB interacts with the five nucleotides immediately upstream of the resume codon, thereby determining the correct selection of the reading frame on the ORF of tmRNA.