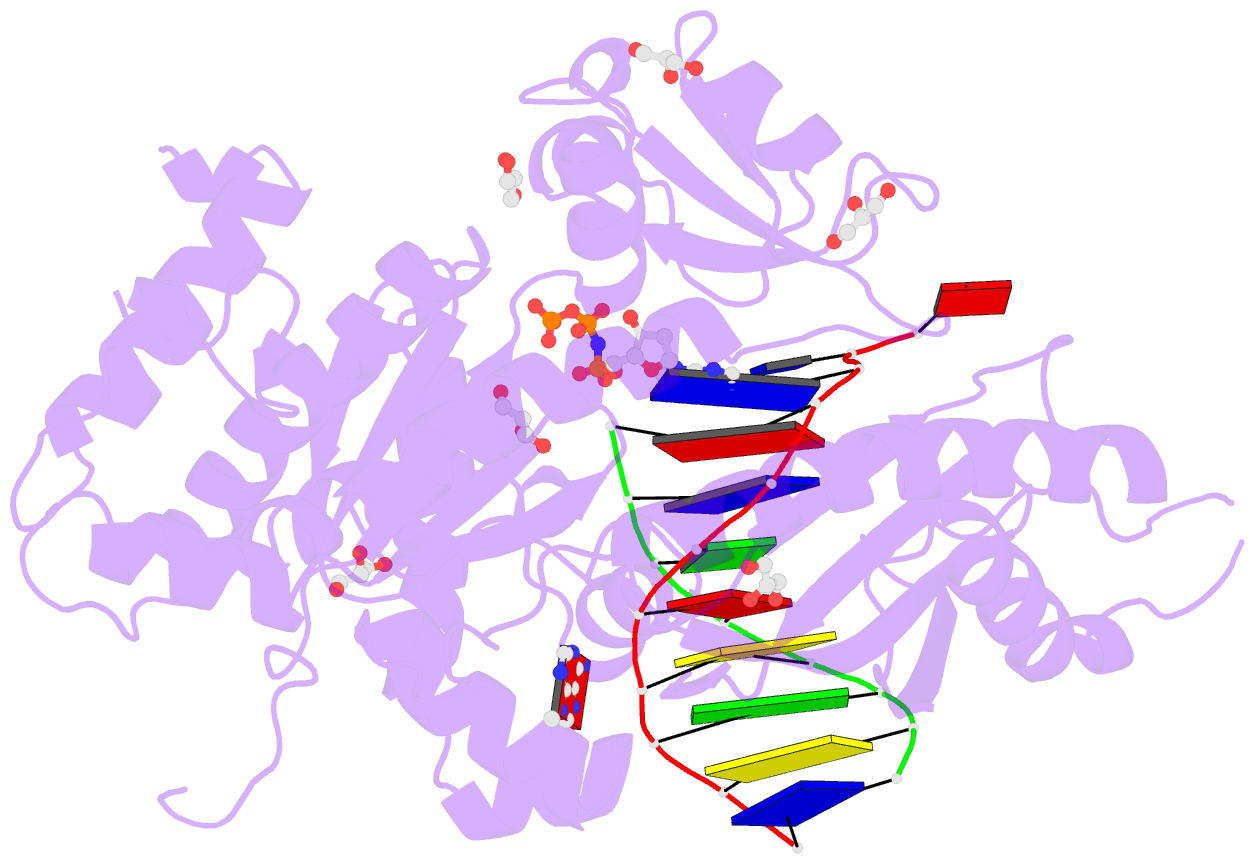
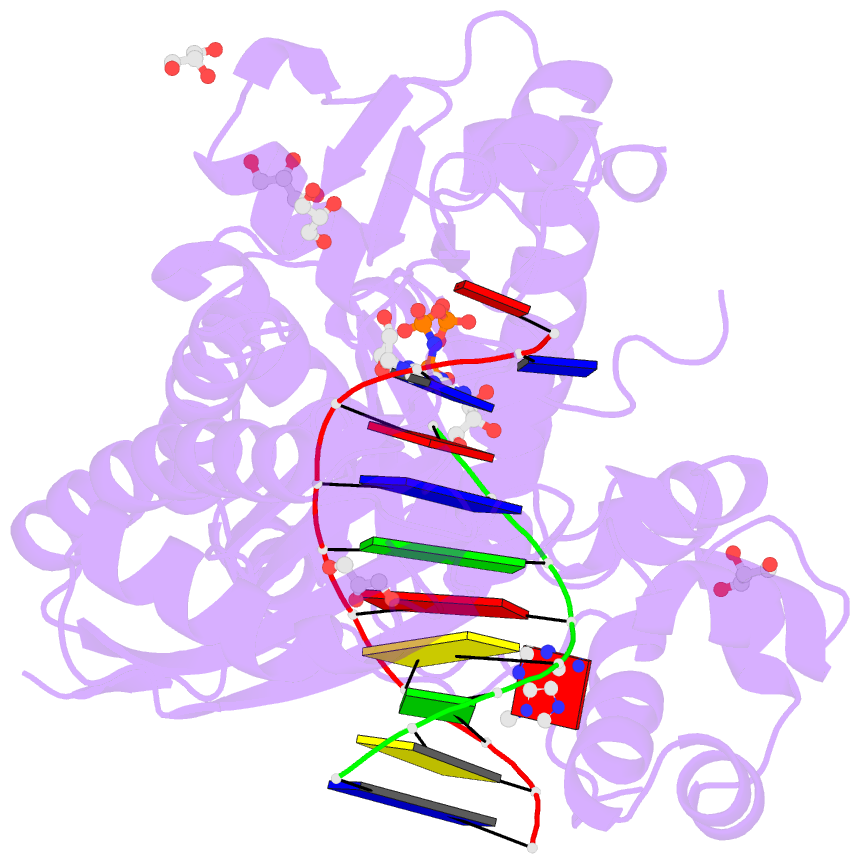
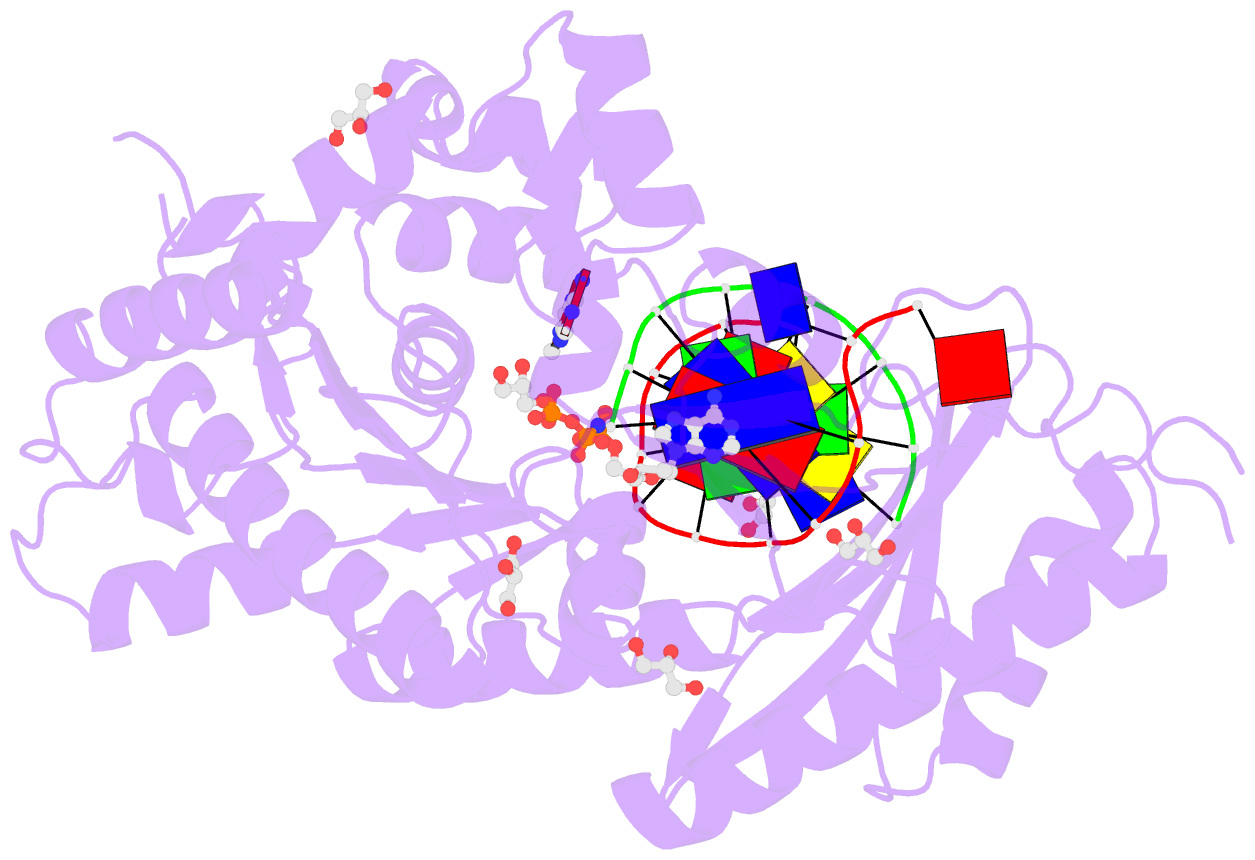
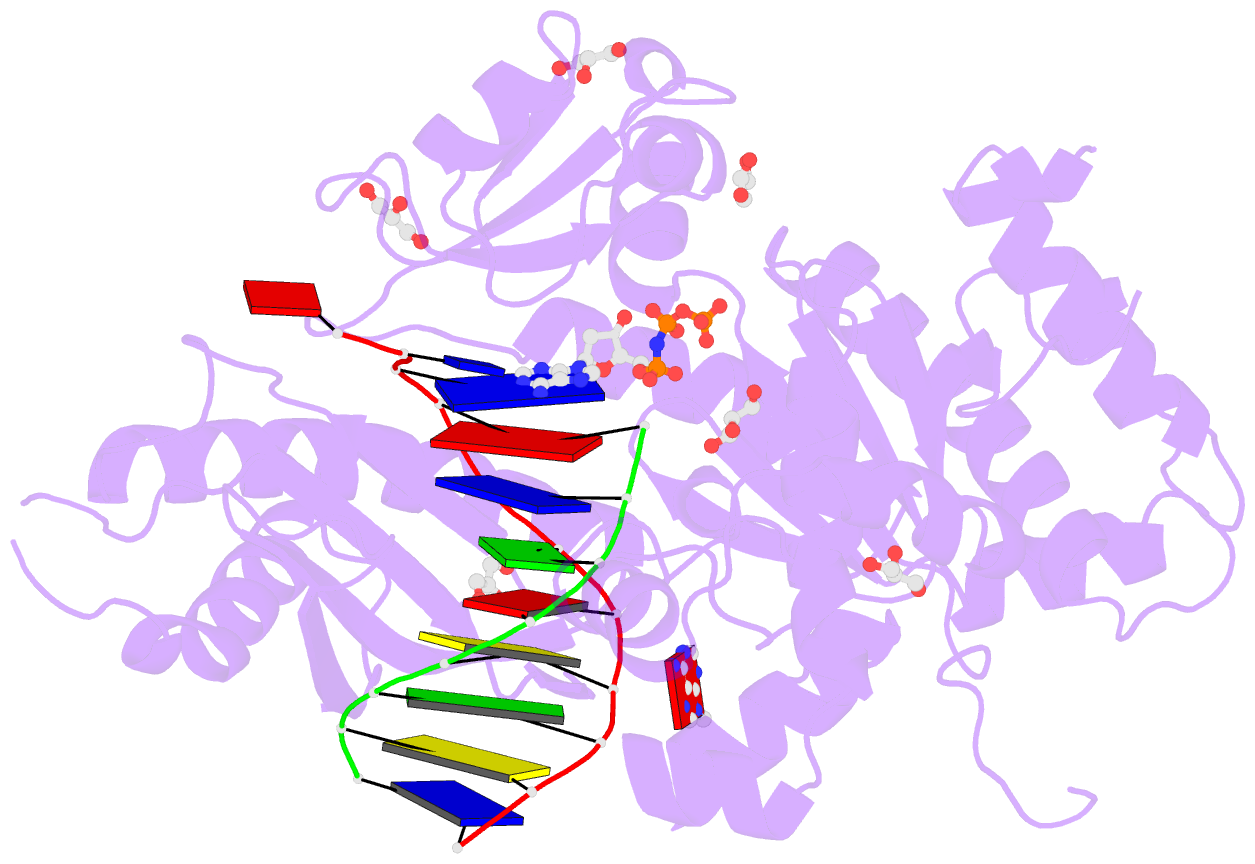
Summary information and primary citation
- PDB-id
- 3jaa; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- cryo-EM (22.0 Å)
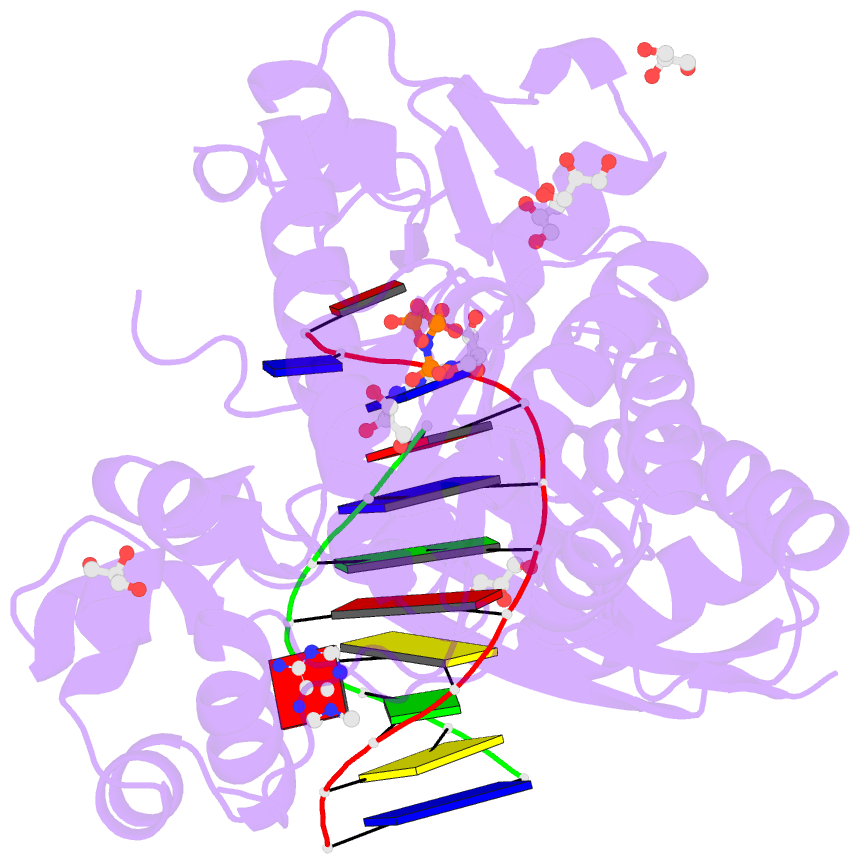
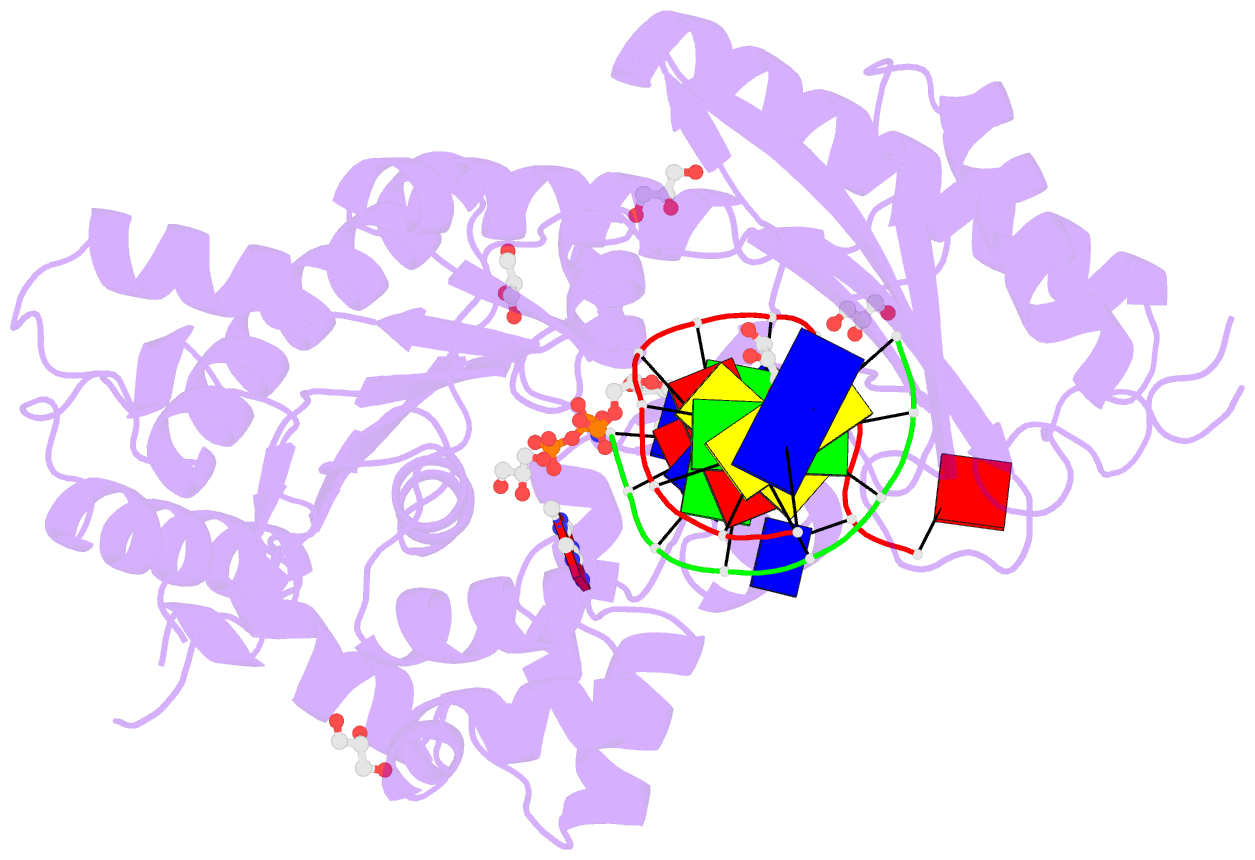
- Summary
- Human DNA polymerase eta in complex with normal DNA and inco nucleotide (nrm)
- Reference
- Lau WC, Li Y, Zhang Q, Huen MS (2015): "Molecular architecture of the Ub-PCNA/Pol eta complex bound to DNA." Sci Rep, 5, 15759. doi: 10.1038/srep15759.
- Abstract
- Translesion synthesis (TLS) is the mechanism by which DNA polymerases replicate through unrepaired DNA lesions. TLS is activated by monoubiquitination of the homotrimeric proliferating cell nuclear antigen (PCNA) at lysine-164, followed by the switch from replicative to specialized polymerases at DNA damage sites. Pol η belongs to the Y-Family of specialized polymerases that can efficiently bypass UV-induced lesions. Like other members of the Y-Family polymerases, its recruitment to the damaged sites is mediated by the interaction with monoubiquitinated PCNA (Ub-PCNA) via its ubiquitin-binding domain and non-canonical PCNA-interacting motif in the C-terminal region. The structural determinants underlying the direct recognition of Ub-PCNA by Pol η, or Y-Family polymerases in general, remain largely unknown. Here we report a structure of the Ub-PCNA/Pol η complex bound to DNA determined by single-particle electron microscopy (EM). The overall obtained structure resembles that of the editing PCNA/PolB complex. Analysis of the map revealed the conformation of ubiquitin that binds the C-terminal domain of Pol η. Our present study suggests that the Ub-PCNA/Pol η interaction requires the formation of a structured binding interface, which is dictated by the inherent flexibility of Ub-PCNA.