Summary information and primary citation
- PDB-id
- 3jxy; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.5 Å)
- Summary
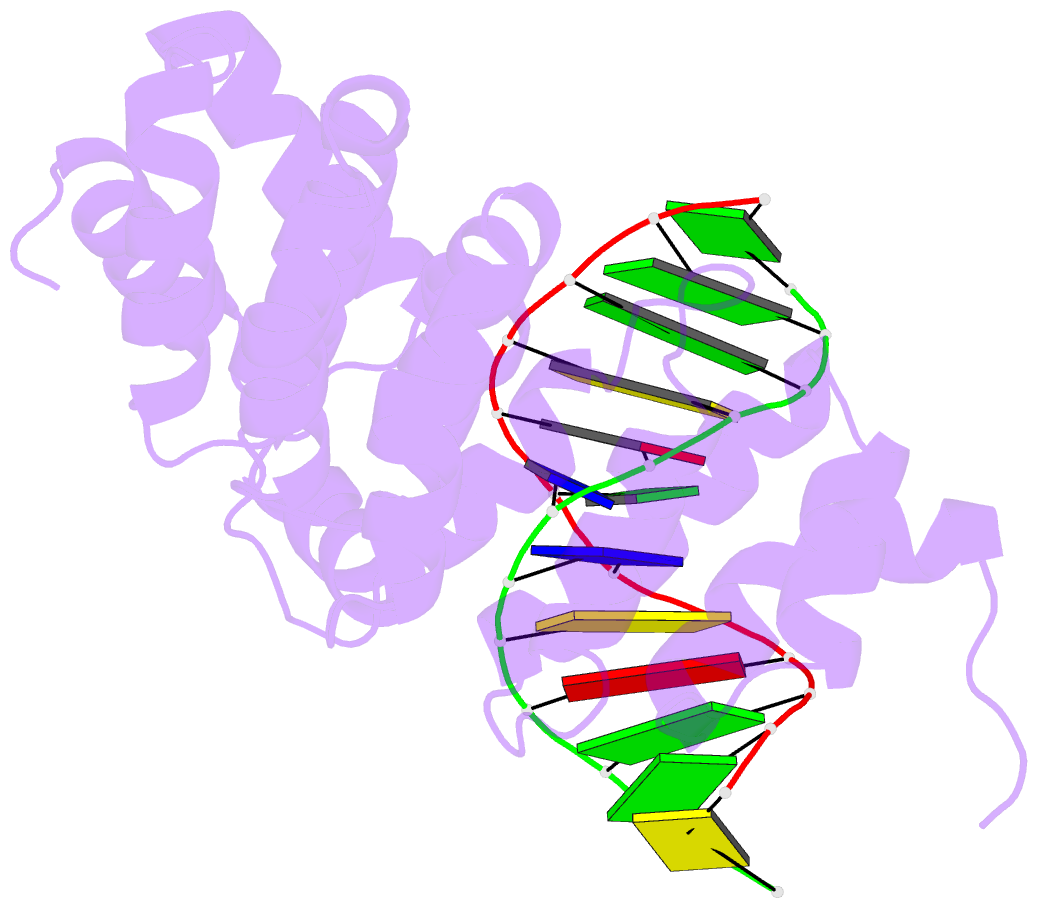
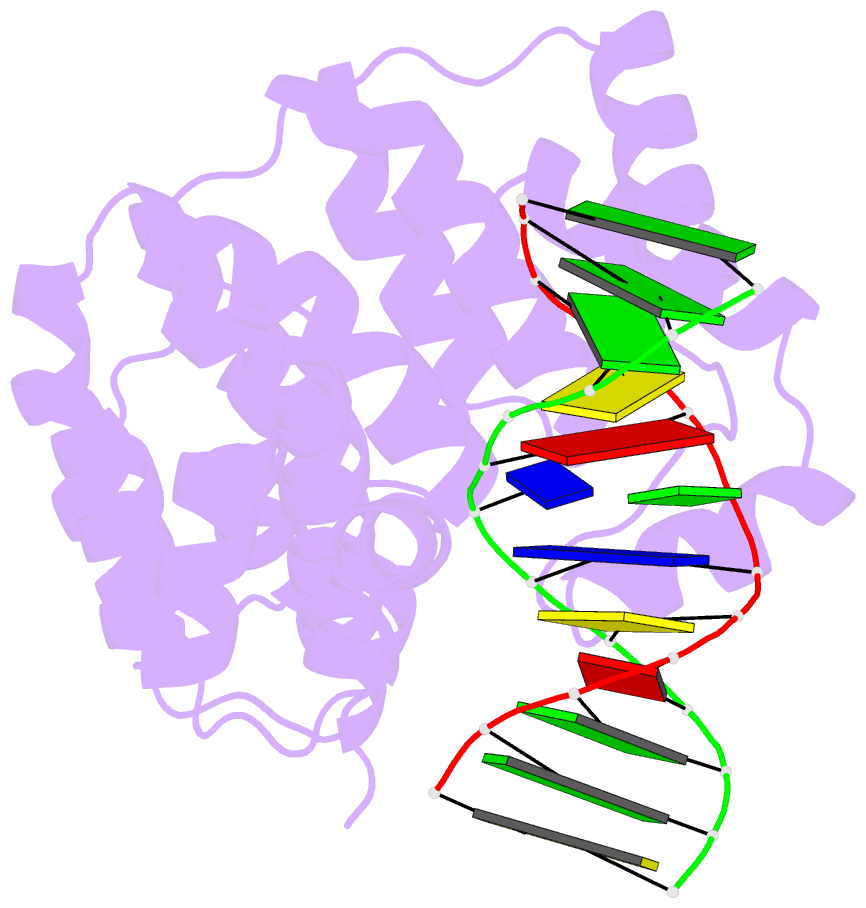
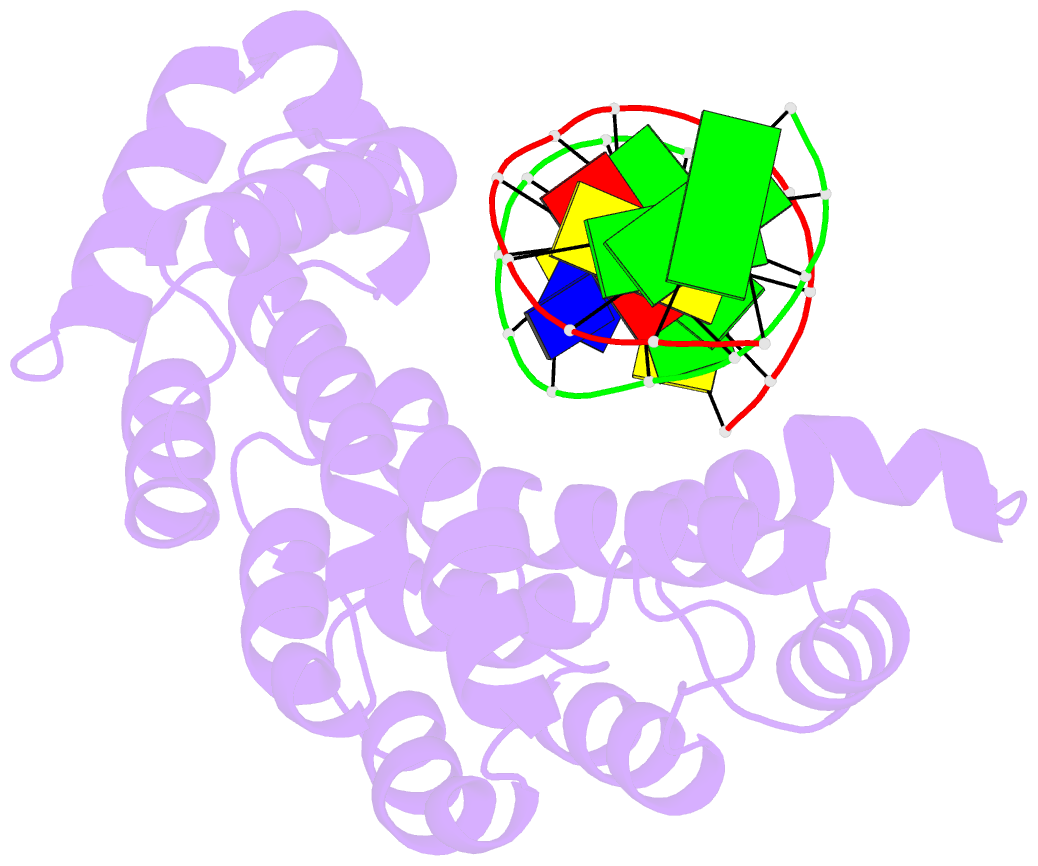
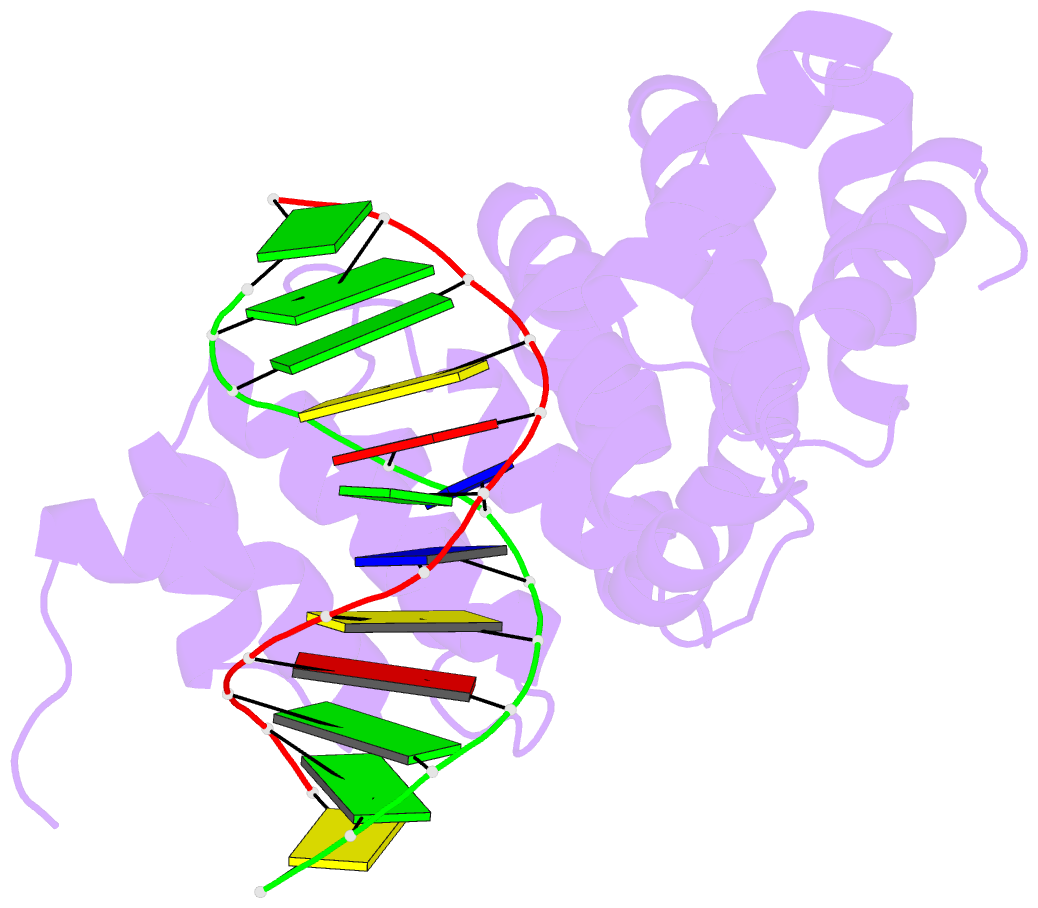
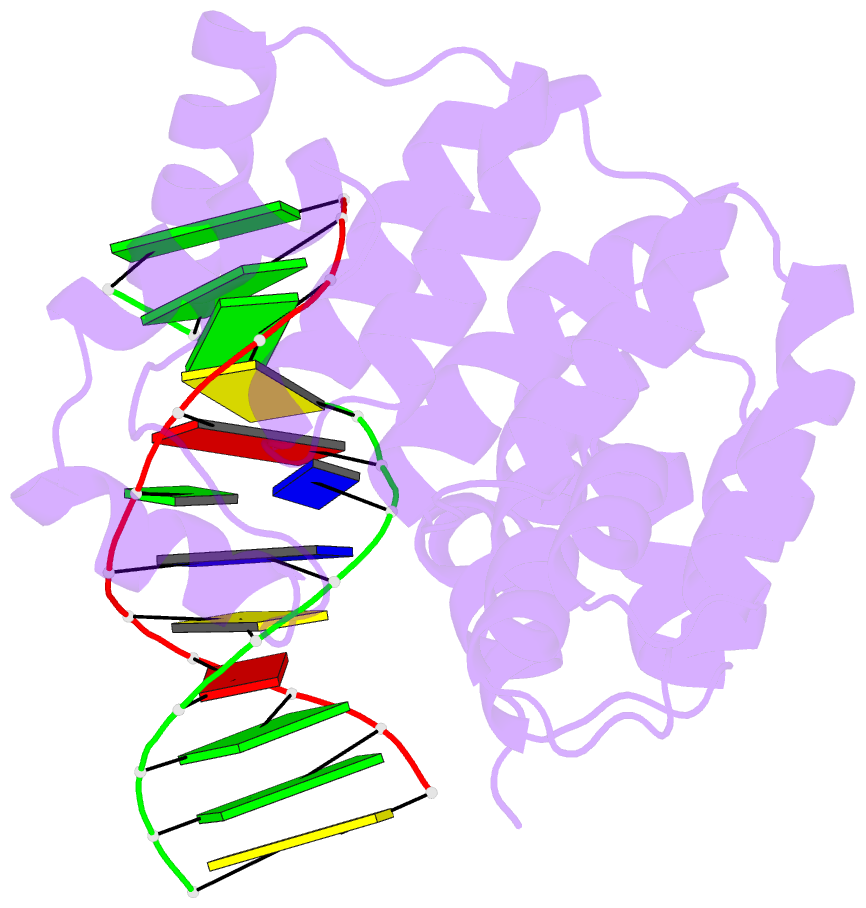
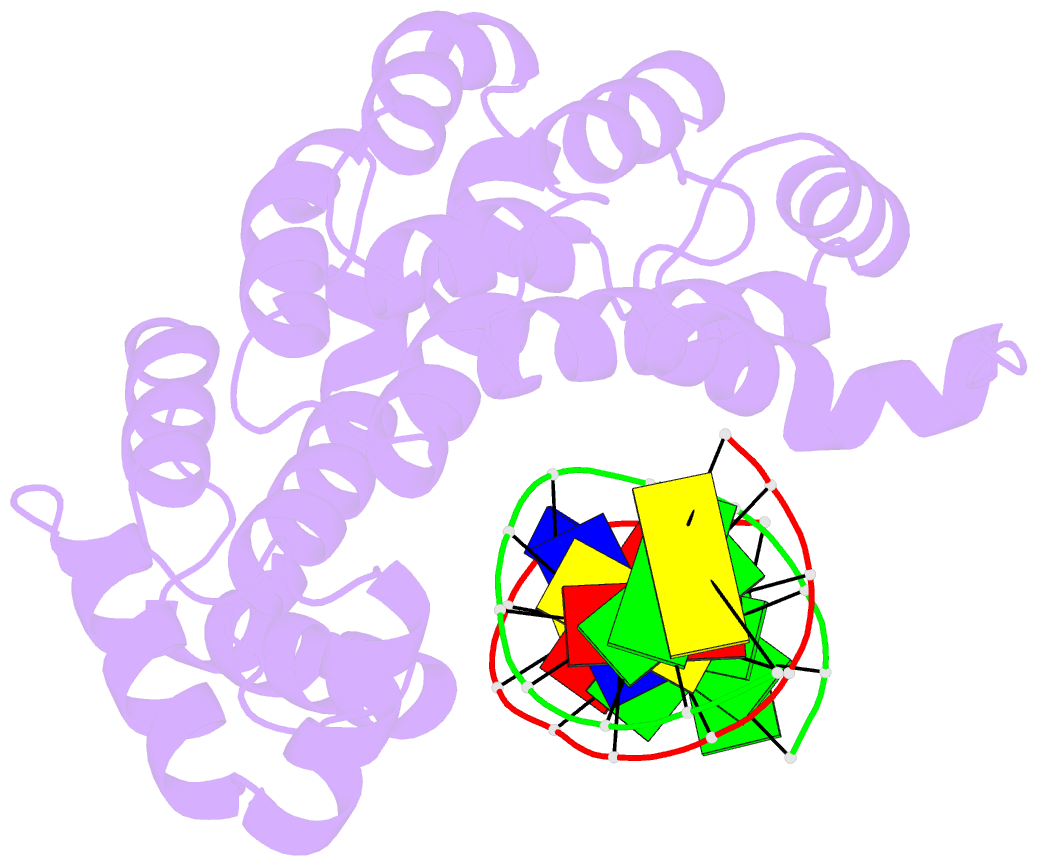
- Bacillus cereus alkylpurine DNA glycosylase alkd bound to DNA containing a gt mismatch
- Reference
- Rubinson EH, Gowda AS, Spratt TE, Gold B, Eichman BF (2010): "An unprecedented nucleic acid capture mechanism for excision of DNA damage." Nature, 468, 406-411. doi: 10.1038/nature09428.
- Abstract
- DNA glycosylases that remove alkylated and deaminated purine nucleobases are essential DNA repair enzymes that protect the genome, and at the same time confound cancer alkylation therapy, by excising cytotoxic N3-methyladenine bases formed by DNA-targeting anticancer compounds. The basis for glycosylase specificity towards N3- and N7-alkylpurines is believed to result from intrinsic instability of the modified bases and not from direct enzyme functional group chemistry. Here we present crystal structures of the recently discovered Bacillus cereus AlkD glycosylase in complex with DNAs containing alkylated, mismatched and abasic nucleotides. Unlike other glycosylases, AlkD captures the extrahelical lesion in a solvent-exposed orientation, providing an illustration for how hydrolysis of N3- and N7-alkylated bases may be facilitated by increased lifetime out of the DNA helix. The structures and supporting biochemical analysis of base flipping and catalysis reveal how the HEAT repeats of AlkD distort the DNA backbone to detect non-Watson-Crick base pairs without duplex intercalation.