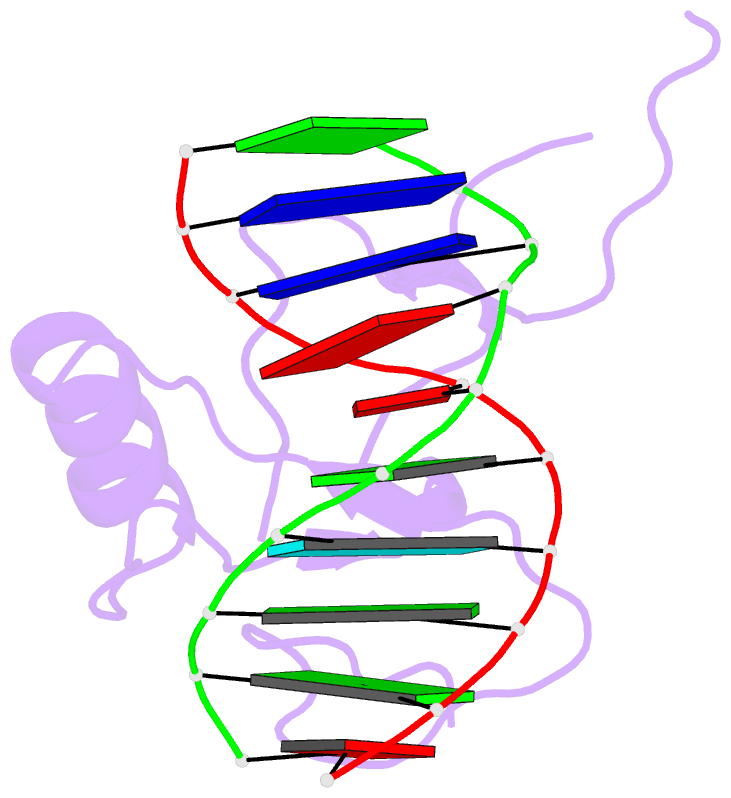
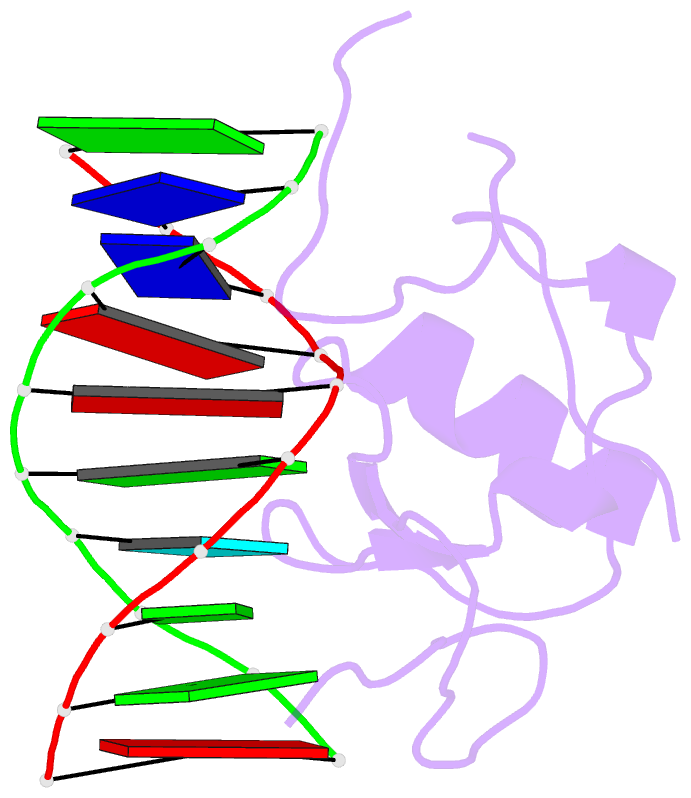
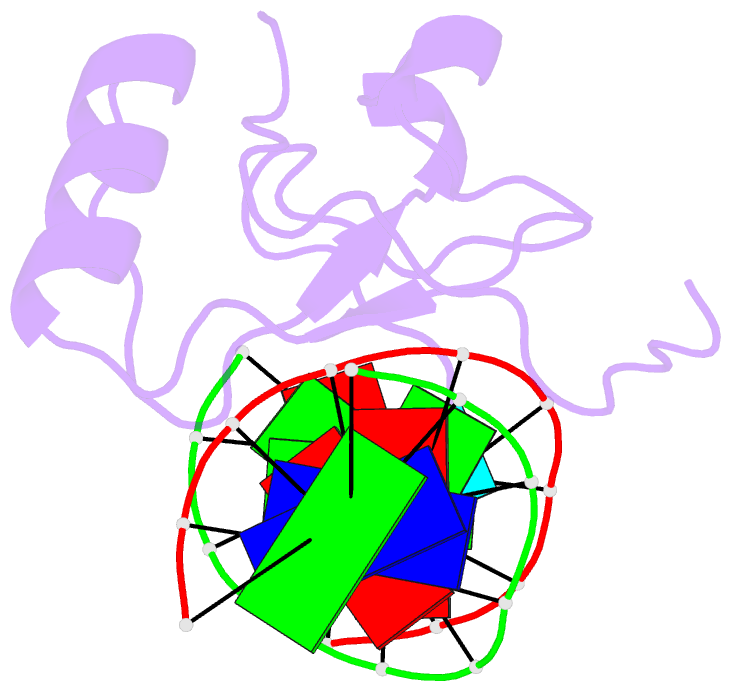
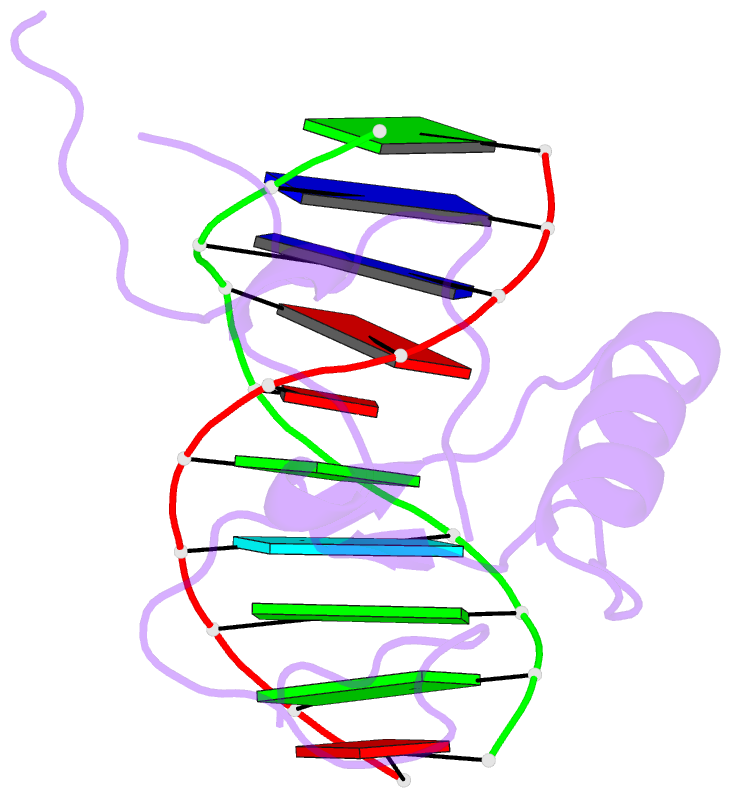
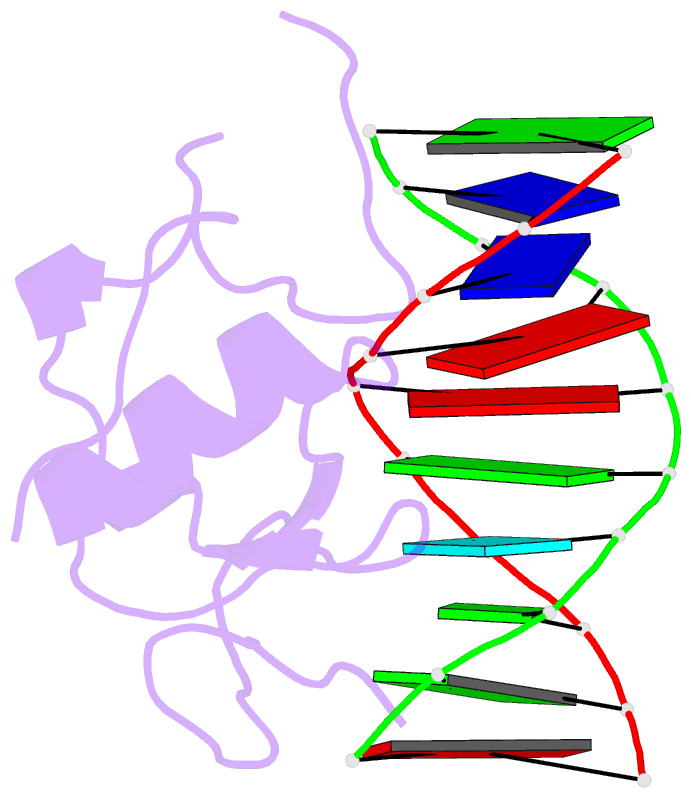
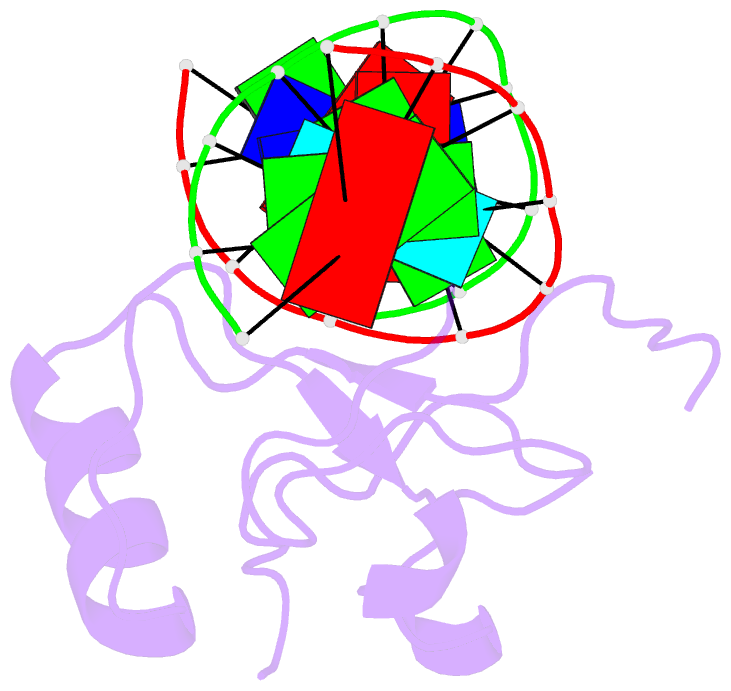
Summary information and primary citation
- PDB-id
- 3kde; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (1.74 Å)
- Summary
- Crystal structure of the thap domain from d. melanogaster p-element transposase in complex with its natural DNA binding site
- Reference
- Sabogal A, Lyubimov AY, Corn JE, Berger JM, Rio DC (2010): "THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves." Nat.Struct.Mol.Biol., 17, 117-123. doi: 10.1038/nsmb.1742.
- Abstract
- THAP-family C(2)CH zinc-coordinating DNA-binding proteins function in diverse eukaryotic cellular processes, such as transposition, transcriptional repression, stem-cell pluripotency, angiogenesis and neurological function. To determine the molecular basis for sequence-specific DNA recognition by THAP proteins, we solved the crystal structure of the Drosophila melanogaster P element transposase THAP domain (DmTHAP) in complex with a natural 10-base-pair site. In contrast to C(2)H(2) zinc fingers, DmTHAP docks a conserved beta-sheet into the major groove and a basic C-terminal loop into the adjacent minor groove. We confirmed specific protein-DNA interactions by mutagenesis and DNA-binding assays. Sequence analysis of natural and in vitro-selected binding sites suggests that several THAPs (DmTHAP and human THAP1 and THAP9) recognize a bipartite TXXGGGX(A/T) consensus motif; homology suggests THAP proteins bind DNA through a bipartite interaction. These findings reveal the conserved mechanisms by which THAP-family proteins engage specific chromosomal target elements.