Summary information and primary citation
- PDB-id
- 3kmd; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.15 Å)
- Summary
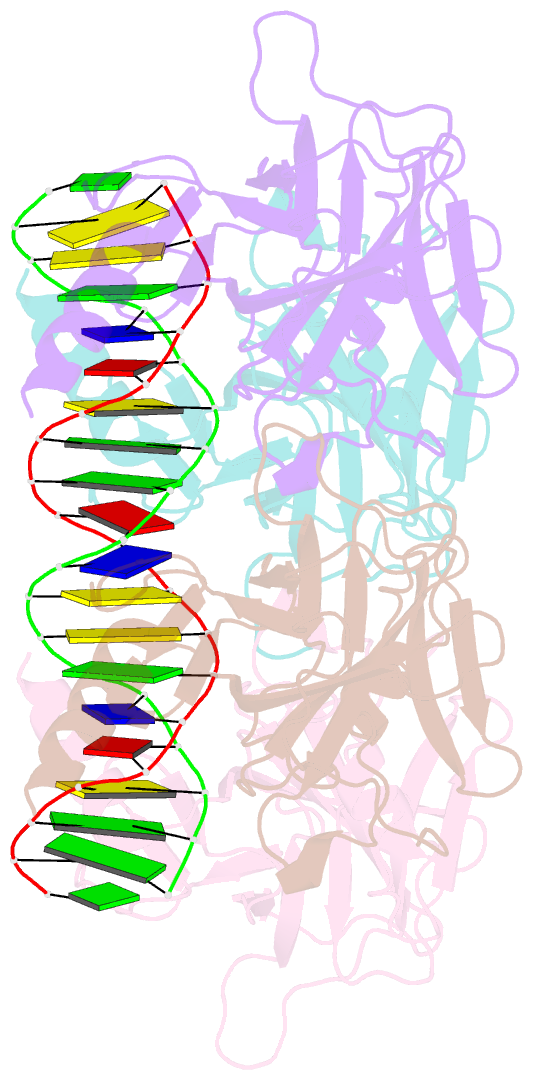
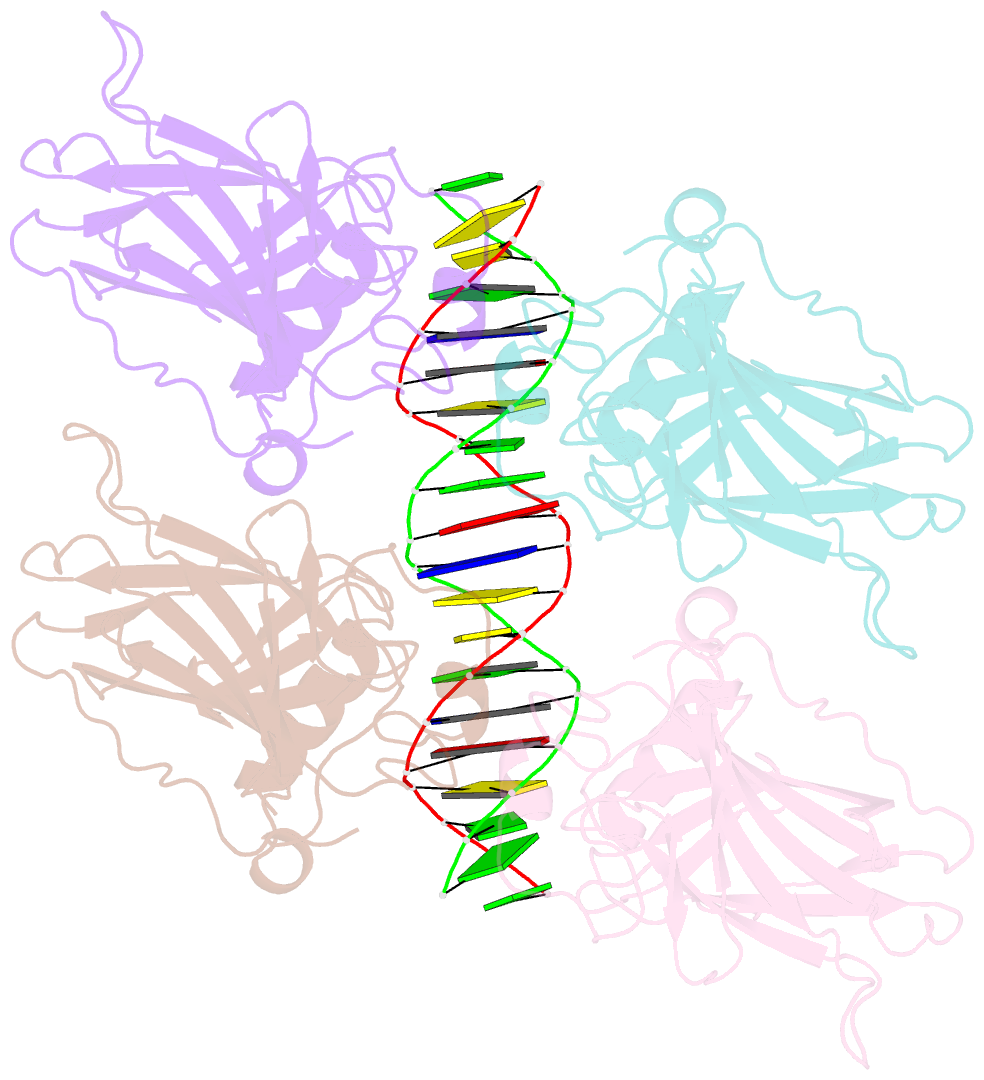
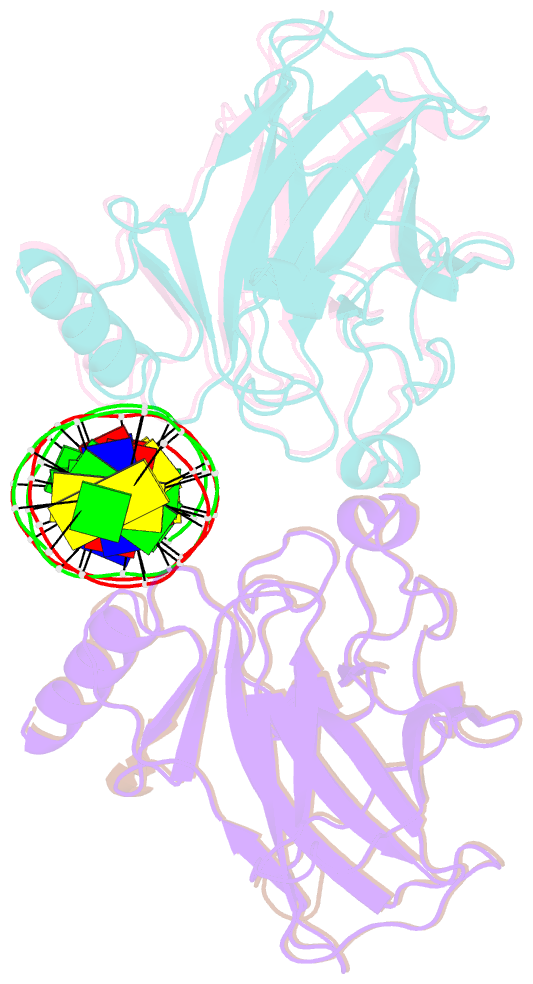
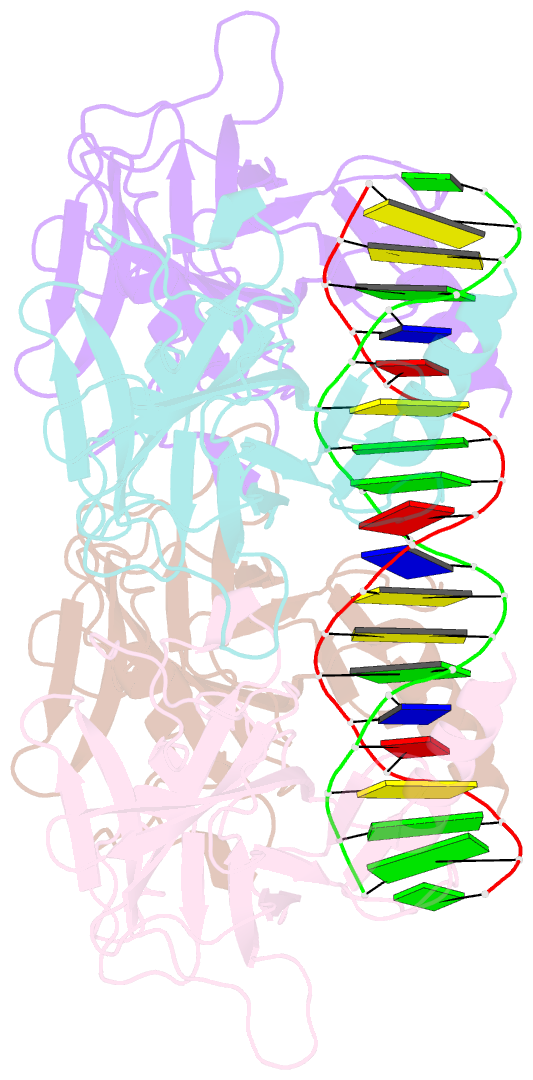
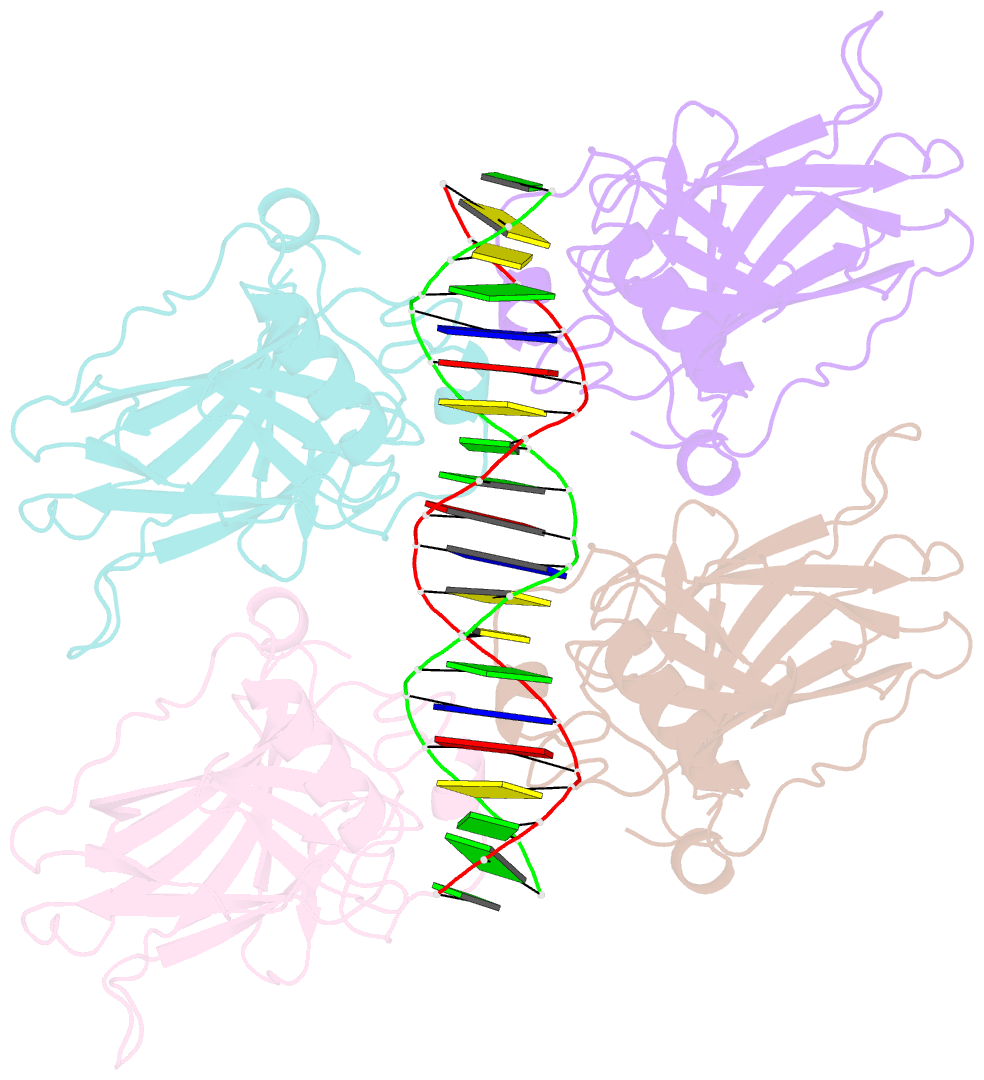
- Crystal structure of the p53 core domain bound to a full consensus site as a self-assembled tetramer
- Reference
- Chen Y, Dey R, Chen L (2010): "Crystal structure of the p53 core domain bound to a full consensus site as a self-assembled tetramer." Structure, 18, 246-256. doi: 10.1016/j.str.2009.11.011.
- Abstract
- Recent studies suggest that p53 binds predominantly to consensus sites composed of two decameric half-sites with zero spacing in vivo. Here we report the crystal structure of the p53 core domain bound to a full consensus site as a tetramer at 2.13A resolution. Comparison with previously reported structures of p53 dimer:DNA complexes and a chemically trapped p53 tetramer:DNA complex reveals that DNA binding by the p53 core domain is a cooperative self-assembling process accompanied by structural changes of the p53 dimer and DNA. Each p53 monomer interacts with its two neighboring subunits through two different protein-protein interfaces. The DNA is largely B-form and shows no discernible bend, but the central base-pairs between the two half-sites display a significant slide. The extensive protein-protein and protein-DNA interactions explain the high cooperativity and kinetic stability of p53 binding to contiguous decameric sites and the conservation of such binding-site configuration in vivo.