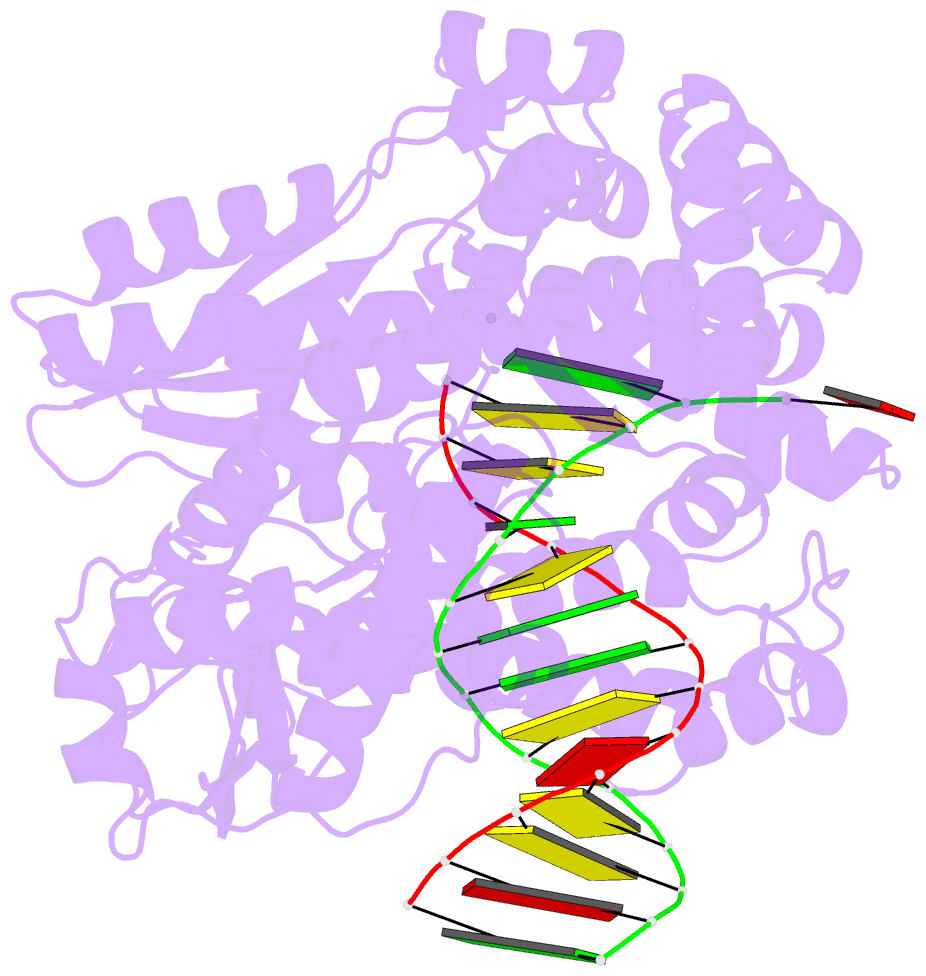
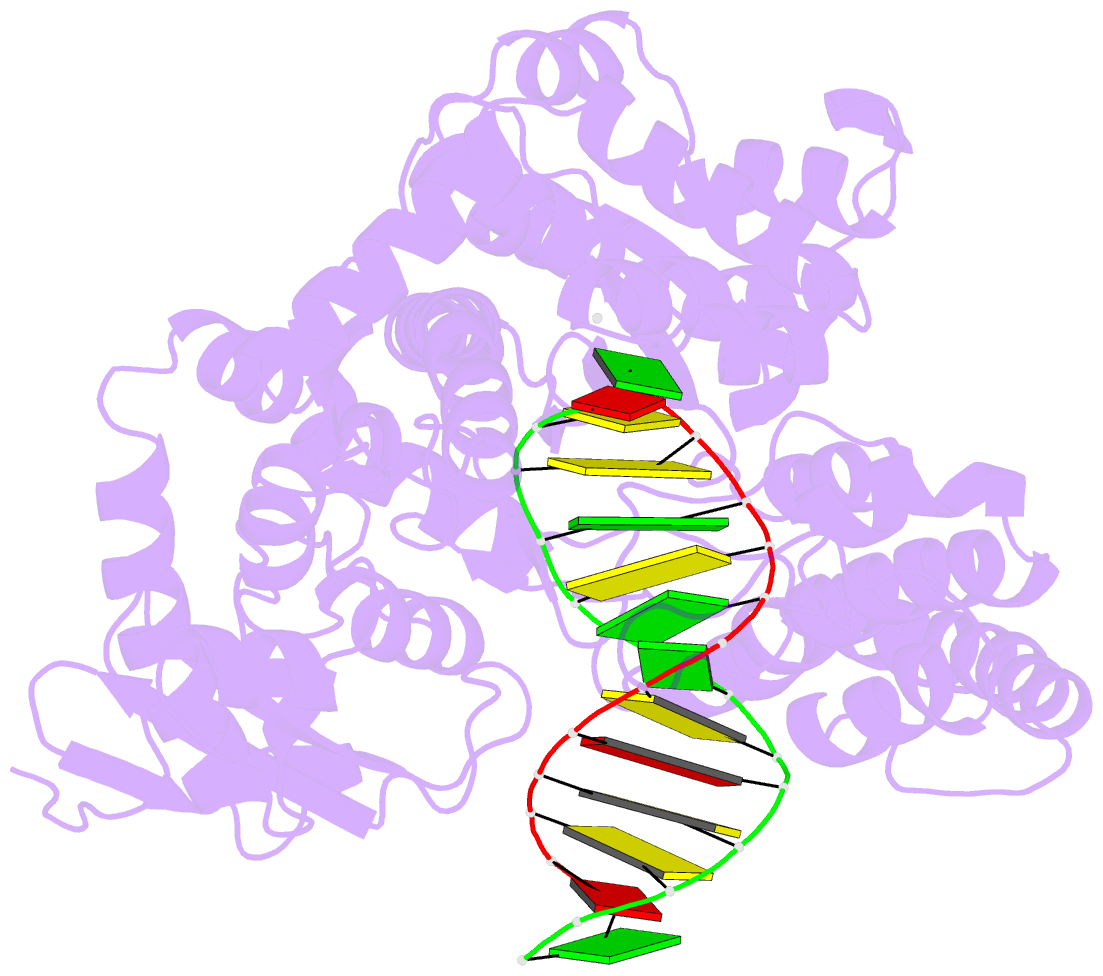
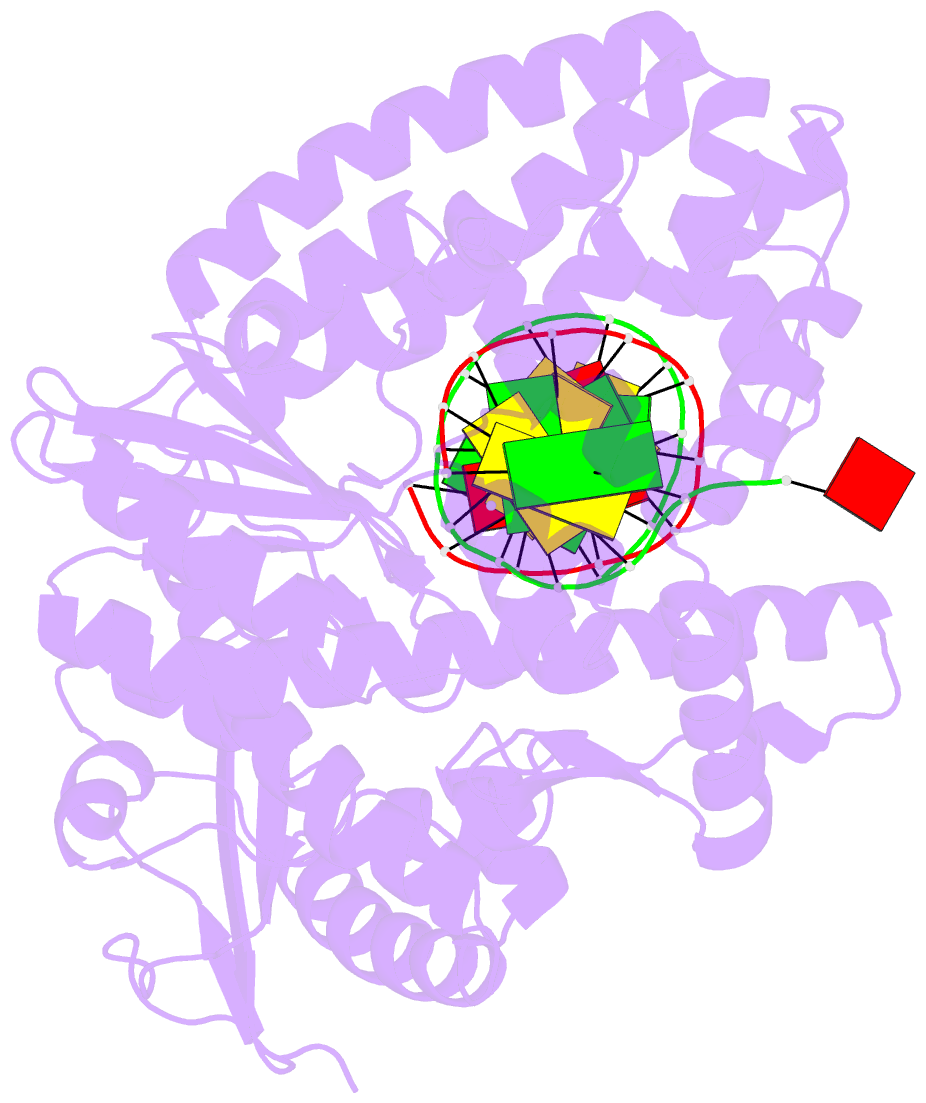
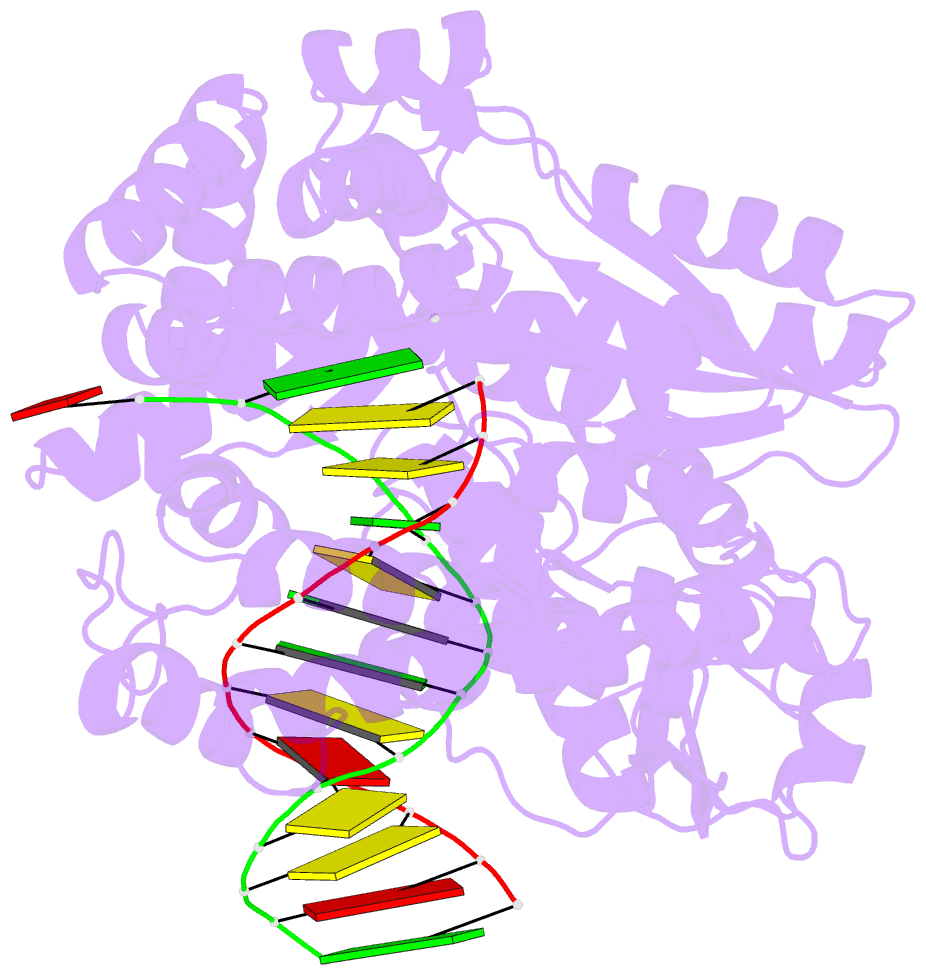
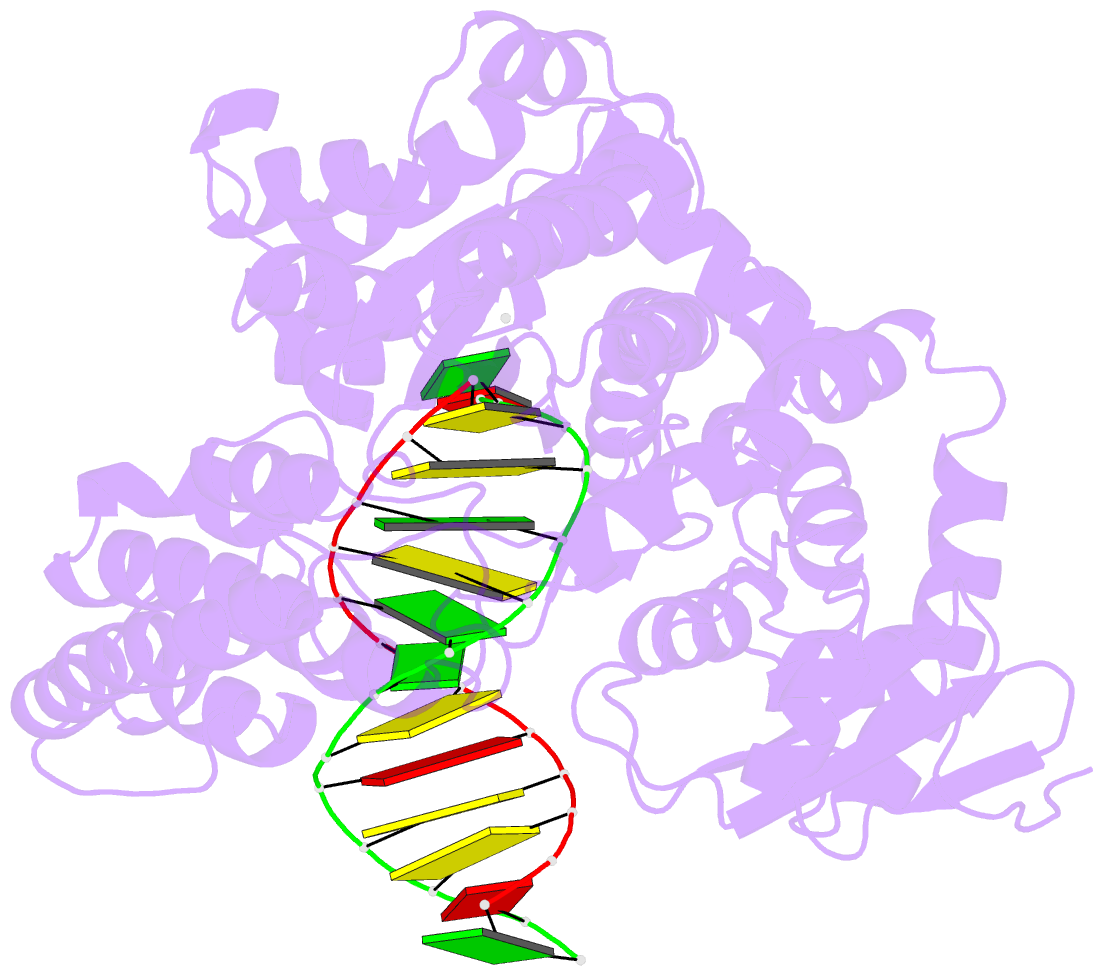
Summary information and primary citation
- PDB-id
- 3ktq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.3 Å)
- Summary
- Crystal structure of an active ternary complex of the large fragment of DNA polymerase i from thermus aquaticus
- Reference
- Li Y, Korolev S, Waksman G (1998): "Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation." EMBO J., 17, 7514-7525. doi: 10.1093/emboj/17.24.7514.
- Abstract
- The crystal structures of two ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I (Klentaq1) with a primer/template DNA and dideoxycytidine triphosphate, and that of a binary complex of the same enzyme with a primer/template DNA, were determined to a resolution of 2.3, 2.3 and 2.5 A, respectively. One ternary complex structure differs markedly from the two other structures by a large reorientation of the tip of the fingers domain. This structure, designated 'closed', represents the ternary polymerase complex caught in the act of incorporating a nucleotide. In the two other structures, the tip of the fingers domain is rotated outward by 46 degrees ('open') in an orientation similar to that of the apo form of Klentaq1. These structures provide the first direct evidence in DNA polymerase I enzymes of a large conformational change responsible for assembling an active ternary complex.